Living bacteria write pictures and movies to DNA
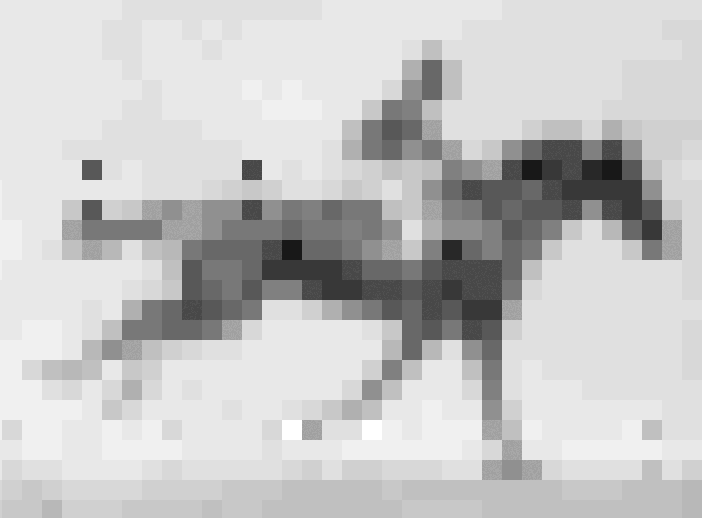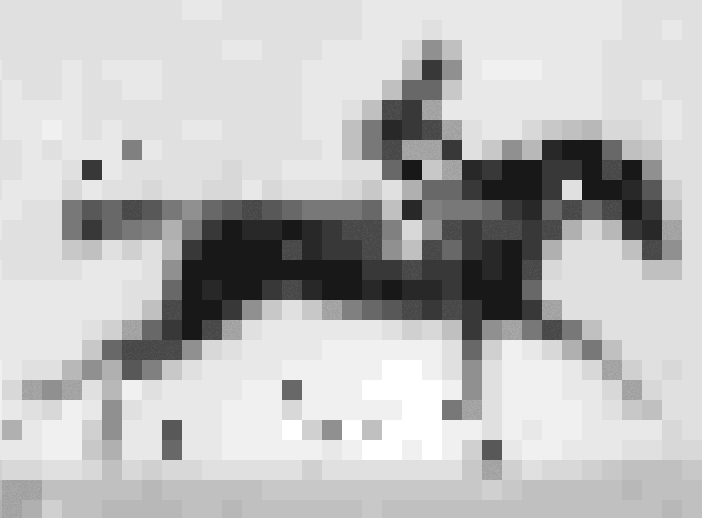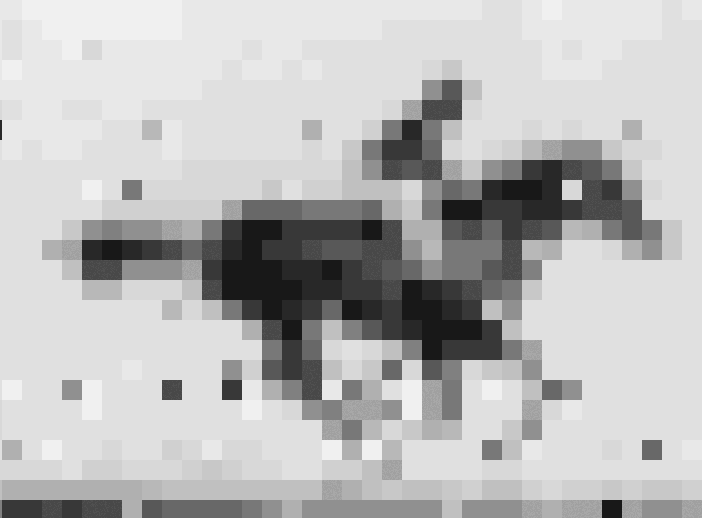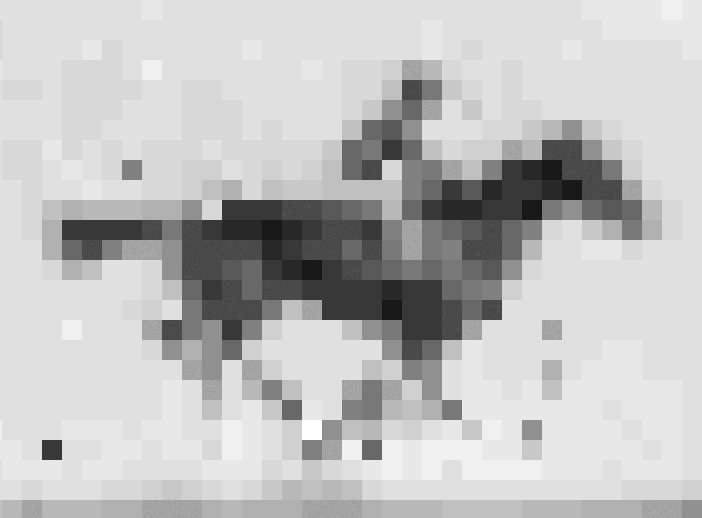
Crispr–Cas9 tool used to store information in the genome of living E. coli
In recent years, data as diverse as Shakespeare’s sonnets, Martin Luther King Jr’s ‘I have a dream’ speech and an Amazon.com gift card have all been stored as DNA. Now, scientists in the US have taken the idea one step further, inserting pictures and movies into the genomes of living bacteria.
