Blowing up brain tissue with swelling polymer delivers sharper images
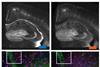
Tissue samples inflated to four times their size to overcome microscopy limitations

Tissue samples inflated to four times their size to overcome microscopy limitations