With drug-resistant bacteria constantly in the news, what is being done to develop better treatments? Phillip Broadwith takes a look
Modern medicine is facing a significant threat. Our antibiotics are becoming less and less effective, and we aren’t developing new ones nearly fast enough to keep up. Hardly a few months go by without someone foretelling the return to a time when seemingly minor infections become life-threatening incidents.
So what is the problem? Why is antibiotic development progressing so slowly? Put simply, the research is very difficult, and the economics are not entirely favourable. ‘When we create a new antibiotic that works well against resistant pathogens, the world will say thank you, then put it in reserve until they absolutely have to use it,’ says David Payne, head of antibacterial research at GlaxoSmithKline (GSK). ‘That’s absolutely appropriate and as it should be, but the return on investment in that scenario is challenging for a drug company.’ Antibiotics are as expensive to develop as other drugs, but if their use is restricted to small populations for short treatments, then recouping those costs is trickier.
GSK is one of the few large pharmaceutical companies still pursuing antibiotics research, but Payne admits that his unit makes up a relatively small part of the company. Instead of a large in-house research programme, GSK has turned to public–private partnerships to share some of the financial and scientific burdens. As a partner in Europe’s Innovative Medicines Initiative, the company is working with academic and industrial collaborators to address some of the challenges in developing new drugs against resistant bacteria.
GSK has also joined forces with the US government’s Biomedical Advanced Research and Development Authority, which is providing up to $200 million (£128 million) in funding over five years. The structure of this partnership is as significant as the money, says Payne. Unlike many other agreements, it is not focused on a single candidate compound, but allows resources to be moved between projects. This means that if one molecule fails – as often happens in drug development – the research can continue without renegotiating the contract.
Target in sight
GSK currently has three antibacterial compounds in the early stages of clinical trials, Payne says, each focused on a different route to beating resistance. One is a new variant of the well-established cephalosporin family. The second inhibits type 2 topoisomerase, which is the enzyme that unwinds DNA as it is replicated. This is the same target as existing fluoroquinolone antibiotics, but the drug binds at a different site. ‘That means the mutations that make bacteria resistant to fluoroquinolones won’t confer the same resistance against these compounds,’ Payne explains.
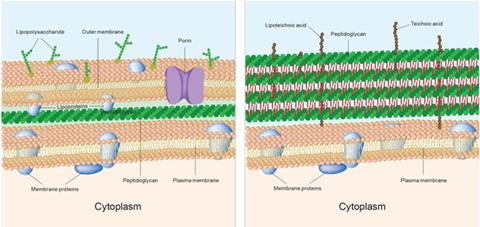
The third is perhaps the most interesting – it inhibits polypeptide deformylase, which is an entirely new mechanism of action. Payne describes the hunt for new ways to kill bacteria as ‘one of the most significant fundamental challenges we face’. This is particularly true for Gram negative bacteria, which have extra layers of protective cell walls that prevent drugs getting into the cells, and have evolved efflux pump proteins to push them back out again if they do manage to penetrate the cell wall.
There’s nothing magical about natural products; they’re still just small molecules
David Spring
The hunt for new modes of action is long and difficult. David Spring from the University of Cambridge in the UK thinks that while those factors make it unattractive to industry, it is an area where academic researchers can make useful contributions. Spring argues that one reason why companies aren’t finding many new drugs is that they’re looking in the same places all the time. ‘Most programmes start with a high-throughput screen of the company’s proprietary compound library,’ he says. ‘If you look at those libraries, a lot of people have been making the same kinds of molecules – there isn’t a great deal of skeletal diversity.’
Spring also points to the fact that many of the most successful antibiotics are derived from natural products – often produced by other organisms as a defence against infection. ‘There’s nothing magical about natural products,’ he says; ‘they’re still just small molecules.’ But what natural product-derived drugs do tend to have is more 3D complexity: for example, stereocentres or macrocyclic and polycyclic scaffolds. ‘We try to make libraries of skeletally diverse, 3D molecules that are natural product-like, then we screen them for antibacterial activity. If there’s even a tiny hint of activity, you can then start to investigate and optimise that lead using more traditional medicinal chemistry.’
One bug, one drug
As a side effect, exploring new areas of ‘chemical space’ like this means that any leads that emerge are unlikely to look anything like existing drugs. This makes the intellectual property around them more valuable and patents easier to establish and defend. It also tends to make the molecules more selective – either for killing bacteria without harming the patient’s cells, or for killing just pathogenic bacteria, leaving the ‘good bacteria’ populations on our skin and in our guts intact.
This is the kind of treatment that Cambridge-based start-up Discuva is aiming to develop. ‘We’ve become used to using relatively broad spectrum antibiotics, and having generic drugs that are very cheap,’ says Discuva’s chief executive, David Williams. ‘But the broad spectrum drugs don’t work any more in Gram negative infections.’ That wipes out the generics market, and provides an incentive for developing innovative drugs. ‘This is an ideal opportunity to redefine the way we treat microbial infection,’ he says.
Instead of looking for new broad spectrum drugs, which by their nature will be very susceptible to resistance, Williams argues we should be developing more targeted therapies for specific infections. These could then be mixed and matched in combinations to help prevent resistance developing by increasing the number of hits bacteria have to take to survive the treatment.
Discuva has developed screening technology that allows it to elucidate a potential antibiotic’s mode of action, as well as any potential resistance mechanisms. It works by screening an active lead compound against enormous libraries of mutant strains of the relevant type of bacteria, with a high density of mutations spanning the entire genome. Looking at which of the mutants survive best gives a picture of what proteins or pathways are essential for the bacterial resistance. This indicates what it could be targeting, but also how easy it is for resistance to arise from random mutations. It also identifies proteins that might confer resistance without being essential to survival, for example efflux pumps or enzymes that rapidly break down the compound.
‘Suddenly you can look inside bacteria and see what’s happening,’ says Williams. Traditional genetic and biochemical methods to get the same kind of information would take years, he adds, whereas Discuva can get the same information in a few days. The process generates vast quantities of data, which is then unpicked using the company’s proprietary bioinformatics platform. That large dataset gives the conclusions a strong statistical backing, so the company can be sure that the results are real and reproducible.
Teaching old drugs new tricks
The flip side of using narrow-spectrum, targeted antibiotics is that you need to know what bug is causing an infection to know what drug to use against it. New diagnostic techniques and equipment are making headway but, in a chicken-and-egg scenario, it will be difficult for them to become established in clinical settings until more drugs that require such specific diagnoses are available.
The drug discovery pipeline is a lottery – chances of success are very small
James Harrison
One possible way to speed such drugs to the market is to reinvestigate older drugs that have fallen out of favour or been abandoned for some reason. ‘Just because a drug is new, doesn’t necessarily mean it’s better,’ says James Harrison, chief executive of Cycle Pharmaceuticals. Also based in Cambridge, Cycle aims to improve access to existing drugs that could be useful but are limited by factors like a lack of suppliers or unsuitable formulations.
Reviving existing drugs is much less risky than trying to discover new ones, both financially and in terms of patient safety, says Harrison. ‘The drug discovery pipeline is a lottery – the chances of success are very small. Some of the drugs we’re looking at have years of safety data behind them.’ The company is working with the US Food and Drug Administration (FDA) to reinstate some of these products via short approval pathways. ‘Unlike most English people, we don’t like waiting in long queues,’ Harrison jokes.
But where do these abandoned drugs come from? Harrison points out that regulatory authorities such as the FDA can approve drugs for marketing, but they can’t force companies to sell products that they decide are not worthwhile for some reason. That means that there are lots of drug files, with all the relevant safety and clinical trial data, sitting collecting dust. ‘A small fraction of these are worth looking at with fresh eyes,’ says Harrison, ‘for example as adjuvants for antibiotics.’
An adjuvant is given along with the antibiotic to enhance its activity. That might be a molecule that helps the drug penetrate the bacterial cell wall or blocks an efflux pump – effectively overcoming the bacteria’s defences and knocking out resistance mechanisms. One way to identify these synergies is by looking for examples where researchers or doctors have tried out combinations successfully, such as in developing countries where they don’t have access to newer, more expensive drugs.
Cycle has identified several potential combination therapies, pairing antibiotics with drugs that were originally approved as treatments for other diseases. By focusing on compounds that are already approved, the company hopes to bypass some of the regulatory hurdles faced by new medicines, and get products onto the market much more quickly. ‘We are always willing to discuss ideas of how to expand the use of existing medicines with researchers or doctors,’ says Harrison.
Call off the troops
No matter how many new treatments we introduce, resistance is an inevitable consequence of fighting against rapidly evolving microbes. In the longer term, we need to understand more about how resistance arises, and perhaps develop strategies to negate, or at least slow down, its development.
Spring is investigating one such strategy: rather than killing the pathogens, they are trying to deter them from attacking their hosts. ‘Resistance arises from the evolutionary pressure to survive in the presence of a drug,’ says Spring. ‘If you allow them to survive, but essentially stop them from behaving badly, the evolutionary pressure is quite different.’
Bacteria communicate using small molecules to allow them to sense how many of their fellows are nearby. This ‘quorum sensing’ controls a variety of functions, for example signalling when to coordinate an attack against the host. Disrupting that communication tips the balance in favour of the immune system, allowing it to clear the infection.
This approach could also help make bacterial populations more susceptible to antibiotics. Spring explains that in some diseases, such as cystic fibrosis, bacteria will form large coordinated networks called biofilms. These giant ‘bacterial cities’ exude protective coatings, express different proteins on their cell surfaces and contain bacterial cells that are often not dividing very rapidly, so drugs that target cell wall growth and division are less effective against them. Breaking down biofilms, or preventing them from forming in the first place, would make the pathogens much more susceptible to antibiotics.
Resisting resistance
As well as how they talk to each other, we still have a lot to learn about how bacteria interact with their environment to maintain their physiological stability, or homeostasis. Stuart Conway, from the University of Oxford, UK, is one of many of researchers investigating this problem as part of the European Network for Integrated Cellular Homeostasis. ‘If you can work out how bacteria maintain homeostasis, you can potentially work out ways to disrupt it and kill them, so you can find new targets for antibiotics,’ Conway says.
Along with collaborators in Aberdeen and California, Conway’s group is looking at a particular ion transport channel called Kef. The channel pumps potassium out of the cell, while hydrogen ions flow in to balance the charge. That lowers the interior pH, which Conway believes helps to protect the cell by protonating nucleophilic groups on essential biomolecules and preventing them from being attacked by electrophilic drugs.
If you can work out how bacteria maintain homeostasis, you can work out ways to disrupt it and kill them
Stuart Conway
This hypothesis is backed up by the fact that the channel opens in response to molecular conjugates of glutathione, which is one of the molecules cells use to mop up reactive electrophiles and tag them for transporting out of the cell. ‘It’s conceivable that this is a kind of synergistic defence mechanism – if your antibiotic contains an electrophile, then it will conjugate with glutathione and be exported from the cell, but at the same time open up this channel to drop the pH and protect DNA from alkylation,’ Conway explains.
Conway’s research is a long way from being translated into a functional drug – or more realistically an adjuvant that could work alongside other antibiotics to block this defence pathway. However, the group has shown that many different glutathione conjugates will activate the channel, and has developed fluorescent probes that can be used to screen for small molecules that bind to it, which is the first stage in looking for potential drugs.
Whether or not the newspaper predictions of a cataclysmic end to the era of antibiotics come to pass, it is clear that chemists, microbiologists and medics have their work cut out. Staying even one step ahead of the bacterial war machine will require attack on every front we can muster, from the immediate firepower of new treatment options, to longer term gains in learning how to strategically outmanoeuvre the bugs.












1 Reader's comment