Turning mature somatic cells back into flexible stem cells using small molecules could revolutionise medicine, especially for regeneration and cancer. Philip Ball reports
Imagine you could take a potion that returns you to an embryonic state, from which you could grow up as someone else. Or another potion that could rearrange your thoughts and features to become another person. Those are just fantasies for humans – but not for our cells. In the past two decades, biologists have become adept at reprogramming mature tissue cells (known as somatic cells) to become another sort entirely, including ones that resemble stem cells with the potential to grow into anything. And whereas the initial work demanded rather drastic addition of new genes, now many of the transformations can be achieved chemically, using mixtures of small molecules. Chemistry alone is proving able to render biology much more fluid and protean than was long thought possible.
These advances could have dramatic implications in tissue engineering and regenerative medicine, perhaps ultimately for treating heart disease or neurological damage. Chemical cell alchemy is also being explored for treating some types of cancer – not by killing the cancer cells but by switching them into other states. ‘We see chemical reprograming [of cells] as the future,’ says cell biologist Saiyong Zhu of Zhejiang University in Hangzhou, China.
Turning back the cellular clock
The story starts two decades ago with the work of Japanese biologist Shinya Yamanaka at the University of Kyoto and his student Kazutoshi Takahashi. Although the word ‘revolution’ is overused in science, what the pair discovered in 2006 surely warrants that term. They showed that mature somatic cells in different tissues of the body could be converted to a stem-cell-like pluripotent state – able to develop into any tissue type. The prevailing view had been that, as the cells in the early embryo become increasingly specialised (the phenomenon called differentiation) during development, the process is one-way: there was no way for cells that have differentiated into, say, skin or liver cells, to find their way back to a pluripotent state. But Yamanaka and Takahashi reported that using viruses to inject into somatic cells a cocktail of just four genes – all known to be active in embryonic stem cells (ESCs) – those cells would revert to a state seemingly like that of ESCs. They called these reprogrammed cells induced pluripotent stem cells (iPSCs). Yamanaka’s pioneering work won him half of the 2012 Nobel prize in medicine or physiology.
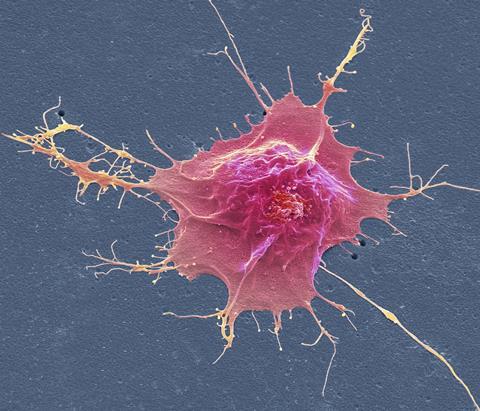
Their initial work used mouse cells, but in 2007 Yamanaka’s team, and independently James Thomson of the University of Wisconsin in Madison, US, reported the same result in human cells. Not only did the discovery overturn conventional wisdom about cell development, but it also suggested exciting medical applications. In principle, iPSCs from an individual could be used to grow new tissues and perhaps even organs to replace ones that are damaged, malfunctioning or worn out. Because they would have that person’s genome, there would be no problem of graft rejection. iPSCs have now been used to grow a wide variety of tissues and miniature organ-like structures, called organoids, in a dish. A further advantage is that, because iPSCs (in contrast to ESCs) are not taken from IVF embryos, they do not fall foul of laws that in some countries restrict or prohibit research with embryonic tissue.
And it’s possible to reprogramme cells not just in a dish but in the body. That could be particularly valuable for regenerating cells and tissues, such as neurons or heart muscle, that can’t easily be grown outside the body and then grafted because they have to be intimately interwoven with those in the existing organs. Researchers are now exploring the uses of iPSCs and cell reprogramming for treating afflictions such as macular degeneration and spinal-cord injury. What’s more, adding genes other than the four now known as Yamanaka factors can transform one mature tissue type directly into another, without reverting first to stem-cell states: say, turning heart connective tissue into heart muscle. These discoveries have added to the growing perception that cell differentiation, rather than unfolding inexorably in a single direction, is much more versatile and ‘plastic’. Maybe, given the right nudges, pretty much any cell could become any other kind of cell.
This is all very exciting, but not without complications. The Yamanaka factors and other reprogramming genes generally have to be carried by viral vectors, which can be awkward to introduce into the body. (Those genes typically encode proteins called transcription factors, which regulate the activity of other genes and induce differentiation by switching them on or off.) Cell biologists subsequently found that reprogramming does not necessarily require extra genes, however. Some of them can be replaced with small synthetic molecules. In 2013, Hongkui Deng of Peking University and colleagues in China reported that they could make mouse iPSCs using no Yamanaka factors at all – the same effect could be induced using seven small molecules. Cells can be reprogrammed by chemistry alone.
Doing that with human somatic cells is more challenging, because our cells have robust mechanisms for resisting such switches of state. But in 2022, Deng and coworkers reported that they had screened libraries of small molecules and found some that, in the right combinations, can induce the switch, for example transforming adult skin cells to a stem-cell-like state. These chemical cocktails contain ingredients that perform a variety of tasks. Some help to disable the ‘epigenetic’ process by which methyl groups are attached to DNA to impose a stable identity for the host cells. Others reactivate Oct4, a key pluripotency gene and one of the Yamanaka factors, which is strongly repressed in human somatic cells. Some of the small molecules have known functions, for example acting as inhibitors of particular enzymes such as kinases, key actors in cell signalling pathways involved in cell proliferation and embryonic development. What others do is still a mystery. The overall goal is to reawaken cell plasticity: to return the cells from a cul de sac to a crossroads.
Chemical regeneration
Using small molecules for reprogramming has many potential advantages. They are easier to make, to standardise, and to introduce to the body, just like ordinary drugs. ‘Small molecules are easy to apply and remove, cell-permeable, more efficient, more cost-effective, and amenable to scale-up,’ says Zhu. So they have more potential to be used not just for in vitro reprogramming but as drugs to stimulate a patient’s cells to repair and regenerate in vivo. And there’s no risk that small molecules, unlike added pieces of DNA, will get integrated into the host cell’s genome.
Isn’t it surprising that small molecules will do these things? It’s one thing for a synthetic drug to bind to the active site of an enzyme and stop it working – but quite another for such a molecule to induce so drastic a biological process as switching a gene on or off, or indeed switching the state of an entire cell. What’s more, there’s no obvious rhyme or reason to the molecules that work. Some of those used by Deng and colleagues were aromatic heterocycles, like pyrimidine derivatives, others have steroid-like polycyclic hydrocarbon frameworks, and so on. These small molecules are typically found by trial-and-error screening rather than logical deduction. This can get results, but it leaves how they work as a black box that can be difficult to prise open.
It seems likely that these small molecules don’t act directly on DNA but work ‘upstream’ of the genes, interacting with the enzymes they encode – for example, changing the behaviour of transcription factors that control other genes. Typically, the state of a cell – what type of cell it is – is determined by interactions of the cell with its environment. For example, molecular or mechanical signals coming from other cells might be received at the cell surface and transmitted via a series of molecular (usually protein) interactions until they affect what happens inside the cell’s nucleus, where the DNA resides in a tangled, fibrous complex with proteins, called chromatin. So the cell’s state is set in a complex, multi-directional negotiation between the genes already active, the networks of molecular interactions within the cell, and the signals from outside. Small molecules that intervene in that dialogue at crucial points can affect the outcome.
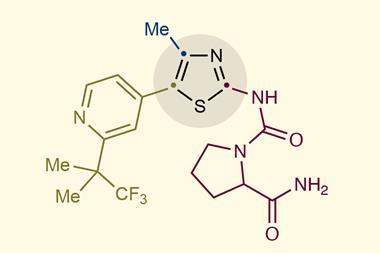
Last year Zhu and colleagues showed how subtle such interventions can be. They investigated how a molecule designated CD3254 (a polycyclic phenol derivative) induces pluripotency in cells. They found that the molecule activates a protein called RXRa, which is involved in a host of biological processes from inflammation to packing of chromatin within the nucleus.
Once activated, RXRa seems to bind to DNA sites that act as ‘promoters’ – rather like on/off switches – for a set of genes encoding proteins involved in the formation of a structure called the RNA exosome, which helps break down RNA molecules that are no longer needed by the cell. So CD3254 induces a cascade of processes: it prompts RXRa to stimulate expression of exosome genes, which then get rid of RNAs transcribed from genes that are thought to maintain the cell state. The researchers kept track of the state of the cells by monitoring the expression of the Oct4 gene that maintain pluripotency. They tethered the Oct4 protein (a transcription factor) to the green fluorescent protein, so that the activity of Oct4 was measured by the brightness of fluorescence.
Since RXRa has other cellular roles too, it wasn’t obvious that a molecule like CD3254 that interacts with it would have a single and predictable consequence. At the moment such efficacious outcomes have to be found by trial and error. ‘The molecular mechanisms of induction signals like CD3254 are very complicated, and our current understanding is still very limited,’ Zhu admits. ‘So the chemical approach to reprogramming is still in its infancy and a lot more studies are required.’
This means that it’s not yet straightforward to design small molecules for reprogramming to order – not least because it’s hard to know what the best binding targets for them are. ‘Currently, the rational design of small-molecule reprogramming agents is still very limited,’ says Zhu. ‘The most effective method is screening and testing.’ But he thinks that AI and machine learning will be increasingly useful for figuring out which molecules might work.
On the other hand, by exerting their effects through the tweaking of natural interactions between molecules in the cell’s biochemical networks, small molecules can be less of a sledgehammer than reprogramming effected by Yamanaka factors and other genes. ‘By triggering innate cellular mechanisms, the chemical approach is much less artificial compared to introducing exogenous transcription factors to abruptly forcing cells to change their identity,’ says Zhu.
These chemical reprogramming methods can suffer from being rather inefficient and taking a long time to transform cell states. But last summer, Zhu and colleagues unveiled a large-scale screening approach that can identify small molecules capable of inducing the switch much faster, comparable with the speed of Yamanaka factors. They hope that such a fast chemical reprogramming technology might be used to give our tissues new capacity for regeneration when they are damaged, and perhaps even to reverse the effects of ageing. ‘Chemical reprogramming methods could be a key approach towards restoring youthful features of cells, tissues and ultimately organisms,’ say cell biologists Sebastian Memczak and Juan Carlos Izpisua Belmonte of the life-sciences company Altos Labs in San Diego, US.
Sending cancer down a different track
The convenience, versatility and relatively gentle effects of small molecules should be particularly valuable for using them as drugs to induce cell transitions in the body. One arena in which these transformations are already becoming clinically useful is in cancer treatment. Cancer chemotherapies are notoriously gruelling, because in general they just destroy cells in the hope that tumour cells will be more affected than healthy ones. But there could be another, gentler way to get rid of cancer cells: by reprogramming them into less destructive or even harmless states.
Every single tumour we’ve looked at has retained plasticity
One of the most striking discoveries in cancer cell biology over the past decade or so is that these cells are not simply proliferating indiscriminately. A given tumour might contain many different types of cancer cell, and there is a logic to how they change that mirrors, in a grotesquely distorted way, the normal developmental process of embryos. Cells of the paediatric brain cancers that Mariella Filbin of the Dana-Farber Cancer Institute in Boston, US, works on are particularly apt to switch between different states. ‘Every single tumour we’ve looked at has retained plasticity,’ she says. ‘When you treat the tumour, the proportions of different states change, or new states come up.’
Perhaps, then, we might find ways of coaxing cancer cells into different, non-malignant states: so-called differentiation therapy. Maybe, says Filbin, ‘you can you push the cells into a particular state that is not plastic and loses a lot of the bad cancer properties such as proliferation and migration, stalling them in a developmental dead end, or that is sensitive to types of therapy that the other cell states were not’.
‘Right now, I’m treating brain tumours that are resistant to chemotherapies and radiotherapies,’ Filbin says. ‘We can’t seem to kill those cells. But maybe we can differentiate them.’
But she adds that the principles governing such cell-state transitions are not very well understood. Much attention has focused on enzymes called chromatin complex inhibitors or degraders. The mammalian chromatin complex called BAF is an assembly of proteins that affects the way chromatin is packaged, and thus how easy it is for the transcription apparatus (such as the molecule RNA polymerase) to get to a gene and express it. BAF seems to hold cells in a plastic state in cancers of brain cells called glia, allowing them to switch back and forth in ways that benefit the tumour, for example to evade chemotherapy drugs. But Filbin and colleagues have shown that using small molecules to interfere with BAF, they can influence how readily the cells transform, and into what. ‘We can make them go back and forth a little bit, though it’s not perfect yet,’ she says.
The researchers used the gene-editing system Crispr/Cas9 to show that a key enzyme in the BAF complex, called BRG1, seems to maintain cells in a glioma tumour in a state that predisposes them to become cancerous. They figured that by disabling BRG1 with a small-molecule inhibitor, they might be able to tip the cells out of their cancerous state. Such molecules are already known and sold by Novartis, and Filbin and colleagues found that two of these could stop glioma cell cultures from proliferating: not killing the cells directly, but stalling their switch to a dangerous state. The two molecules couldn’t work as drugs as they stand, not just because they would needed to be tested for toxicity but because they’re not good at crossing the blood–brain barrier to reach the brain from the bloodstream. All the same, they show that the principle of hitting chromatin remodelling complexes is a good way to control what cancer cells are capable of.
The work illustrates another advantage of this approach: once a good target, such as a chromatin remodelling complex, is identified by screening cancer cells using gene editing methods, small molecules that will bind to them may be already known and commercially available. Often these will already have properties desirable in a drug, such as stability and solubility.
We know from experience that there is hardly ever a single drug that works alone
Filbin thinks that inhibitors of chromatin complexes could be the key target for differentiation therapies. ‘If I had to guess, I think the money will be there.’ However, another approach is called lineage targeting, in which the signalling pathways of a cancer cell are altered to direct it towards a particular lineage or cell type. In this case the usual targets of small molecules are proteins called tyrosine kinases, which are often important in such signalling processes.
These approaches could be used in parallel, using a cocktail of small-molecule reprogramming agents to cajole them into the state you want. ‘We know from experience that there is hardly ever a single drug that works alone,’ Filbin says. ‘Some cells will find ways to get around it. But if you target multiple chromatin complexes at the same time, there’s less chance for that to happen.’
This sort of cancer-cell reprogramming is still at an experimental stage, but already it is being used as a therapy of last resort for some cancers, such as a rare kind of leukaemia called APML and nerve cancers called neuroblastomas that afflict young children. Filbin thinks that paediatric cancers might respond particularly well because they are typically caused by just a few genetic abnormalities, rather than the many often present in adult tumour cells – so there are fewer buttons that need to be pressed.
Chemical reprogramming of cells might thus turn out to be the key for transforming Yamanaka’s remarkable discovery of the reversal of cell fates into a clinically relevant technology. By replacing biological agents with synthetic small molecules, cells might be given a makeover in a way that is milder, more versatile, and easier to use in the body. Perhaps nature doesn’t always know best.
Philip Ball is a science writer based in London, UK

















No comments yet