Researchers have developed a fragment library for drug discovery featuring what they describe as ‘sociable’ three-dimensional fragments. These fragments contain functional groups that make it easier for medicinal chemists to transform initial hits into viable lead compounds. Additionally, the three-dimensionality of the fragments means they can enable exploration of previously underexplored regions of chemical space.
Fragment-based drug discovery gained popularity in the 1990’s and is now an established approach for identifying hit molecules in the early stages of drug development. Fragment libraries have traditionally been populated with flat, two-dimensional molecules. However, there is growing interest in creating libraries of three-dimensional fragments, which offer broader coverage of chemical space and can engage more selectively with protein targets, potentially reducing off-target effects.
Peter O’Brien, from the University of York in the UK, and colleagues have developed three-dimensional fragment libraries in the past. However, they found that many of the selected fragments required bespoke, multistep syntheses and were challenging to elaborate, especially at positions other than the functional groups likely involved in protein binding. Indeed a few years ago, researchers at Astex Pharmaceuticals identified fragment elaboration as a key bottleneck in the transition from fragment hits to lead compounds and developed the concept of fragment sociability.2
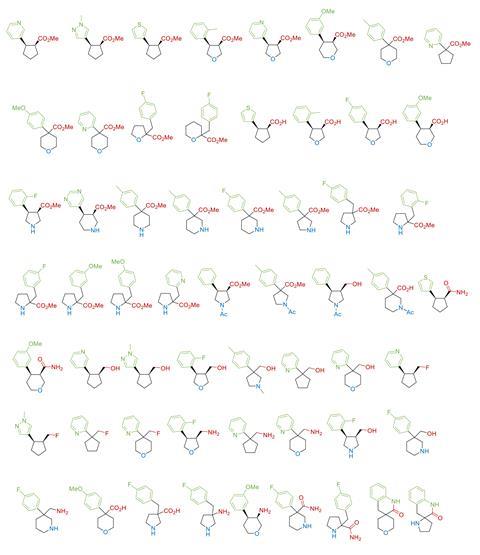
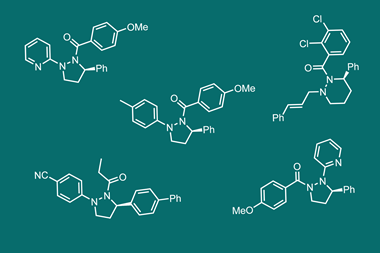
Now, O’Brien working with fellow scientists at York and Astex as well as Pfizer, AstraZeneca, Vernalis, Eli Lilly have designed and synthesised a library of shape-diverse yet sociable 3D fragments. The library comprises 58 fragments, of which 53 are novel structures, each built around a cyclic scaffold such as cyclopentane, pyrrolidine, piperidine, tetrahydrofuran or tetrahydropyran. Each fragment includes an aromatic or heteroaromatic ring to increase the potential for protein interactions, and adheres to the ‘rule-of-three’, to ensure the fragments remain small and soluble enough to bind weakly but specifically to biological targets. To generate the library, the researchers used three synthetic methodologies to generate16 small fragments. By modifying the functional groups of the original fragments, the team synthesised another 42 structurally and chemically diverse 3D fragments.
O’Brien reiterates that issues with fragment libraries usually arise during hit to lead optimisation. He says that collaborating with industry colleagues inspired them to use simple synthetic methodologies so that ‘if we get a fragment hit, we could use that methodology quickly and efficiently to make analogues’.
The researchers screened most of the library against a range of therapeutic targets at the XChem facility at the Diamond Light Source. Fragments from the library were validated in the Sars-CoV-2 main protease and non-structural protein 3, as well as human glycosyltransferase MGATV, an enzyme associated with aggressive metastatic cancers.
Medicinal chemist Mary Wheldon from the University of Dundee, UK, who did their PhD with O’Brien but was not involved with this research, finds the ‘sociability aspect of this work really important’, explaining that often when researchers find a hit, developing it to a lead involves complicated synthesis and this work could allow much quicker hit to lead optimisation.
O’Brien says his team is also developing fragment libraries that combine two-dimensional fragment hits with three-dimensional building blocks – a strategy that has already shown promise for kinase inhibitors.3
References
1 T D Downes et al, Chem. Sci., 2025, DOI: 10.1039/d5sc05819h
2 J D St. Denis et al, RSC Med. Chem., 2021, 12, 321 (DOI: 10.1039/d0md00375a)
3 A R Gomez-Angel et al, J. Am. Chem., 2025, 147, 29292 (DOI: 10.1021/jacs.5c08786)











No comments yet