Assembly theory – an approach that characterises molecular complexity – has been used by scientists in the UK to investigate how selection emerges in chemistry. The method offers new possibilities for making compounds with desired properties in a minimum number of steps based on evolution. ‘By using assembly theory to follow a given path of, say, molecular evolution of a natural product that is a useful drug, we can predict how that product will evolve and literally “jump ahead” in time to get the new molecule rather than waiting,’ says Lee Cronin at the University of Glasgow, UK, who invented assembly theory and led the study.
Chemical space includes every possible compound – all the drugs and materials we already know and those that are still to be discovered – so it contains an almost infinite number of molecules. But many of the potential products may be unstable or synthetically inaccessible, so exploring the vast range of possibilities to find useful molecules can be a huge and challenging task. The method developed by the research team could make things easier.
Assembly theory extracts evolutionary information from the structure. The scientists considered that simple living organisms only need a few hundred different types of special small molecules to function, indicating that the chemical space relevant to biology is only a tiny fraction of the chemical space that is possible. The main issue is then to find out what makes the molecules both thermodynamically possible and biologically accessible.
Molecular family trees
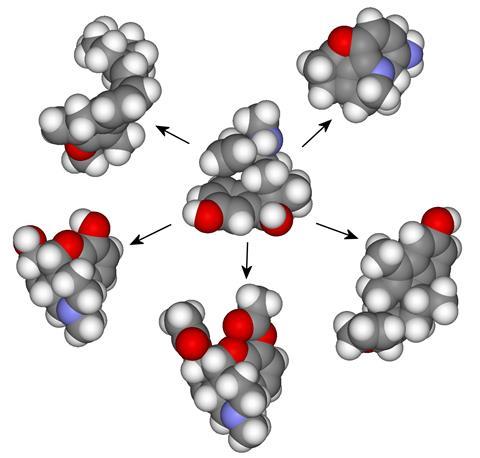
To study this, the scientists created molecular trees by taking different molecules, cutting them into bits and arranging the pieces in order, from smallest to largest, to make the end target. Such molecular trees are like family trees that identify a compound’s ‘parents’ and ‘offspring’. ‘When you take two or more molecules and cut them apart, you can overlay them to find common units and then look to see how you can combine the trees to not only make the two molecules but also novel hybrids,’ explains Cronin.

He adds that the method can be used to combine related molecules but also totally unrelated ones. ‘This approach allows you to find potentially more interesting molecules because you’re accounting for the interesting features that have been selected by evolution and also using the assembly tree to keep to that path rather than getting too lost.’
Alán Aspuru-Guzik, a computational chemist at the University of Toronto in Canada, says that this is a new way of thinking about chemical space. ‘The authors introduce the concept of assembly trees to analyse proximity in chemical space,’ he comments. ‘This methodology seems promising to understand molecular complexity.’
But looking for molecular trees can be a hard mathematical problem, especially as the molecules get bigger, so the researchers developed a Monte Carlo algorithm to help them with the search. ‘Taking simple building blocks, and by connecting or overlapping them with each other, the algorithm proceeds to find the shortest, non-trivial pathways of assembly for given molecules,’ Aspuru-Guzik says. ‘By exploring these pathways in families of molecules, assembly graphs can help explore chemical spaces.’
The team applied the approach to study a class of potent but addictive painkillers – opiates. ‘These molecules were an important target as we’re interested in exploring chemical space for effective new opiates that aren’t addictive and hence so dangerous,’ Cronin says. In the study, nine natural and synthetic compounds were grouped together and broken into assembly pools that could then be used to explore new possible drugs while keeping all the features necessary for the molecules to work.
The authors generated 1000 opiate-like compounds using the assembly tree as well as 1000 random compounds considering the bonds present in opiates. They found that the molecules obtained from the assembly pools showed a much higher similarity to opiates than the random compounds. The products generated using the assembly tree also exhibited similar levels of drug-likeness to opiates, as opposed to random molecules which were less drug-like.
The scientists also demonstrated the potential of the new method in areas such as prebiotic chemistry, genetics and environmental chemistry. ‘It remains to be tested how these new ideas work in a practical design application,’ mentions Aspuru-Guzik. ‘But they can provide a good theoretical framework to think of chemical space.’
References
Y Liu et al, Sci. Adv., 2021, DOI: 10.1126/sciadv.abj2465









No comments yet