Extraordinary crystal structure displays abiotic foldamer with unprecedented complexity
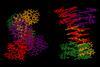
Four aromatic oligoamide helix-turn-helix units assemble in organic solvent into an abiotic architecture with quaternary-like structure
A team surrounding Ivan Huc, from Ludwig-Maximilians-University of Munich in Germany, has made a 12.9kDa pseudo-quaternary structure composed of four aromatic oligoamide helix-turn-helix modules.1 Single crystal x-ray diffraction analysis of the abiotic foldamer shows some previously identified assembly patterns, as well as new ones involving multiple water molecule bridges. ‘This contribution to foldamer research pushes the complexity limit of artificial assemblies,’ remarks Helma Wennemers, a chemical biology expert from ETH Zurich in Switzerland, who was not involved in the project.