Modified arylomycin combines high potency with new mechanism of action
With bacteria becoming resistant to antibiotics and therefore increasingly deadly, US researchers have developed ‘a much-needed new class’ of drug. Medicinal chemists at Genentech in San Francisco, California, have carefully optimised arylomycin antibiotics, found being made by soil bacteria in the early 2000s.
The molecule, G0775, uses a new mechanism of action to kill primarily fiendish Gram-negative bacteria known to resist existing drugs. Consequently the Genentech team, led by senior scientist Christopher Heise claim that it could be an important element in holding back the tide of antimicrobial resistance.
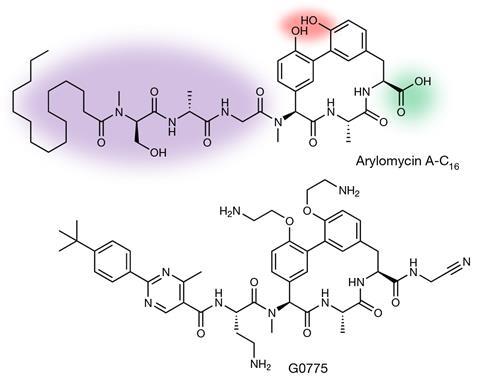
Genentech was drawn to arylomycins, Heise explains, because they bind to and block signal peptidase (SPase) enzymes, which sit on bacteria’s outer cell membranes. Proteins that need to be moved to the outside of the cell membrane are marked with signalling peptides, which the SPases cleave off once they get there, allowing them to fold into their final shape and function properly. That makes them essential to keeping bacteria alive.
As the SPases sit on the cell surface, they were an attractive target for fighting Gram-negative bacteria, which have an extra ‘envelope’ of armour surrounding their cell membrane and preventing drugs getting inside the cell. But initially, the arylomycins were also ineffective against Gram-negative species, owing to a mutation in the SPase enzyme binding site.
The arylomycin scaffold features a tripeptide ring with two phenol groups, a fatty tail on one end, and a carboxylic acid on the other. Genentech kept the tripeptide ring, but changed the fatty tail to make it bind tighter. They also swapped the carboxylic acid with a group that attaches permanently to the SPase, which expanded killing power to otherwise especially hardy bacteria. Modifying the phenol groups also boosted activity, leading to G0775, which kills bacteria at doses a factor of 500 lower than a natural arylomycin, and works in infected mice.
‘The arylomycin natural products were like diamonds in the rough,’ Heise says. ‘We stripped the scaffold down to its core and then systematically polished all of its edges, revealing in the end its potential for remarkable potency.’ Testing G0775 against various bacteria, resistant mutants only emerged rarely, at low drug concentrations. Even then, the antibiotic still killed mutant bacteria after increasing doses 4- to 16-fold.
Heise’s team has recently put Genentech’s ‘first small molecule antibiotic targeting multi-drug resistant Gram-negative bacteria’ into clinical development. They remain ‘actively interested in the arylomycin scaffold and molecules like G0775, ultimately with the hope of delivering a new medicine to patients,’ Heise says.
The University of East Anglia’s Laura Carro-Santos calls G0775’s development ‘fantastic news’, highlighting both its potency and lack of toxicity. She hopes that it will both help understand the new SPase bacterial target and revitalise ‘the dwindling antibacterial pipeline’.
References
P A Smith et al, Nature, 2018, DOI: 10.1038/s41586-018-0483-6











No comments yet