A new approach to fragment-based drug design
Researchers in the US and Germany have developed a metallofragment library for fragment-based drug discovery. As the first of its kind, this library could help medicinal chemists access drug leads with underexplored three-dimensional shapes that are more likely to interact with biological targets.
Fragment libraries enable medicinal chemists to identify core molecular structures that act as starting points for new drugs. Traditionally, fragment libraries consist of one-dimensional and two-dimensional organic fragments. A lack of three-dimensional fragments in these libraries means that medicinal chemists could fail to find drugs occupying three-dimensional chemical space. Now, Seth Cohen, Nils Metzler-Nolte and their groups have developed a metallofragment library to help change that.
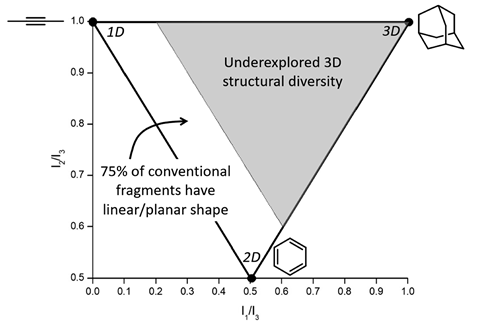
The team’s library contains 71 small coordination compounds. Each compound has a single metal ion and they are all synthetically accessible from readily available starting materials. These fragments serve as an unreactive scaffold for building more complex drug molecules. In its entirety, the library consists of thirteen classes of fragment with members of each class sharing the same metal and core geometry, with differing functional groups and heterocycles. The fragments used include an assortment of various sandwich, half-sandwich and octahedral complexes, demonstrating the diverse topology of the library. And although it has fewer fragments than existing libraries, it occupies a greater proportion of three-dimensional space.
‘Protein active sites are three-dimensional spaces. This suggests that molecules with greater intrinsic three-dimensionality should be able to better exploit this target space,’ explains Cohen ‘Fundamentally, we also simply don’t know what we are missing by not exploring more three-dimensional fragments. Exploring this space may reveal unexpected opportunities and insights.’
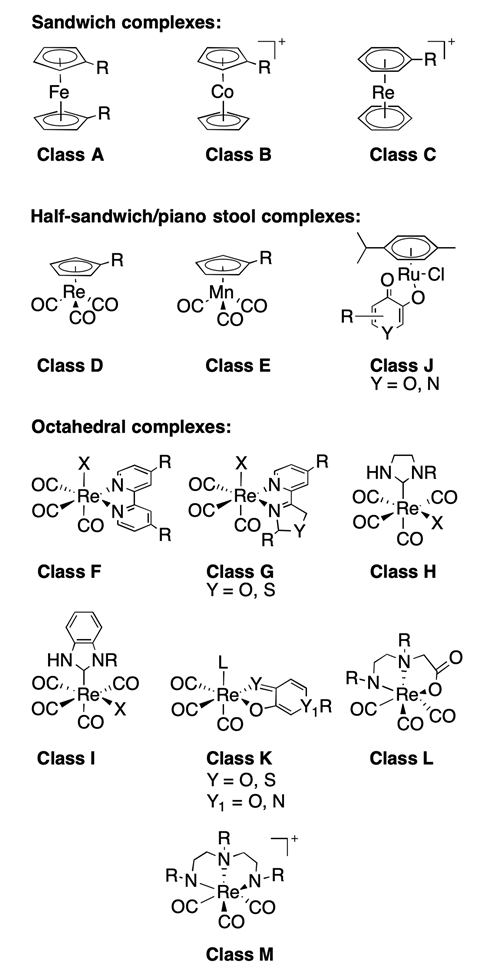
The team assessed the effectiveness of the metallofragment library by screening the fragments against three therapeutic targets to gauge their potential in antibacterial, antiviral and anticancer drug designs. Those three targets originated from New Delhi metallo-β-lactamase-1, an enzyme that confers antibiotic resistance on bacteria; the polymerase complex that replicates the viral RNA genome of influenza A; and heat shock protein 90, which is overexpressed in some tumours. They found that the library matched or outperformed traditional screening libraries in these areas, highlighting the usefulness of these fragments as a basis for future drug design.
Medicinal chemist Derek Lowe finds the work interesting and says the library could uncover some interesting three-dimensional chemotypes. However, he questions if industrial researchers will really use metallofragment libraries. ‘I think it faces an uphill fight into the mainstream of fragment-based drug discovery. It’s true that there are organometallic compounds used in therapy and diagnostics, but that doesn’t mean that they’re first choices, by any means. I believe that most industrial drug discovery researchers will not wish to take on the added complications and risk of developing them.’
Cohen acknowledges there are barriers to using metals in medicine. The team is now developing examples of hit-to lead compounds based on metallofragments to convince the medicinal chemistry community of the usefulness of their drug development strategy.







1 Reader's comment