For the first time, small strings of amino acids have been used to encode enormous chemical libraries of over 40,000 compounds. Thanks to these peptide libraries, researchers were able to identify a number of drug candidates for cancer.
The concept of chemical libraries is popular in drug discovery. ‘It relies on two key components – encoding and decoding information,’ explains joint first author Nathalie Grob from the Massachusetts Institute of Technology in the US. It works like barcodes: after attaching unique ‘tags’ to a range of small molecules, researchers investigate their affinity to proteins previously associated with known diseases. Then, the best binders are identified by simply reading the tag. Usually, chemists tag compounds with DNA fragments, which enables quick and simple separation and interrogation of the library. But DNA limits the possibilities, mostly because of the sensitivity and incompatibility of nucleotides with certain chemicals and catalysts – such as strong acids, transition metals, even radicals.
Peptides, however, can tolerate a far greater range of conditions, including cross couplings making them an excellent alternative to DNA. On top of this improved reaction compatibility, tagging with peptides can accelerate the decoding process as well, explains Grob. ‘We decode the tags using mass spectrometry, whereas [DNA tags] require enzymatic reactions and amplification methods like PCR,’ she says.
Christian Heinis, an expert in peptides at EPFL in Switzerland, says this technique has ‘great advantage[s] for the construction of chemically and structurally diverse libraries’. This comes, mostly, from the robust resistance of protected peptides to harsh chemical conditions, which allows the production of many different molecules and building blocks. ‘To my knowledge, it’s the first time that peptides [work] for encoding and screening small molecule libraries,’ he adds. ‘Moreover, the solid phase synthesis [method] could allow an automated production.’
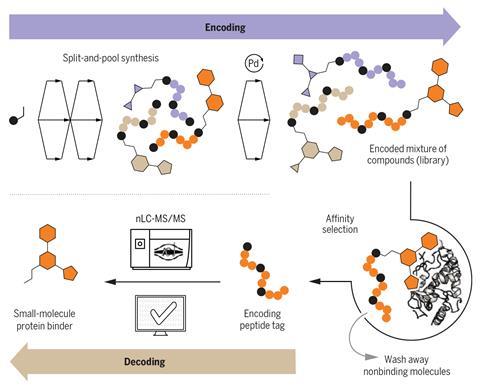
However, the process is still manual. ‘Nevertheless, we synthesise full libraries in less than a week,’ says joint first author Simon Rössler. ‘Solid phase isn’t like solution synthesis, our optimised reactions are really fast and don’t require purifications,’ he explains. They follow what’s known as a splitting-pool strategy in combinatorial chemistry. The synthesis of the library forms a ‘family-tree’ branched structure with many shared steps. ‘We spend a significant amount of time weighing the reagents, rather than synthesising the peptides,’ he jokes.
Nina Hartrampf, a peptide and organic chemistry expert based at the University of Zurich, Switzerland, explains that the starting molecule for the library is immobilised in a resin with a branched linker that connects both the amino acid barcode and the small molecule. ‘The reactions on both ends are then performed iteratively,’ she says. ‘To generate a library … the resin is divided into smaller portions, which are then treated differently. Each portion is functionalised with different small molecule fragments, then encoded with the respective amino acids. ‘After each pair of reactions – functionalisation and encoding – the entire resin is recombined and split again, to perform the next set; this is how the sequence space is generated.’ Thanks to the synthesis on solid support and the high reaction yields, reaction outcomes are clean without purification. ‘The challenge really comes with the [peptides’] encoding and decoding,’ she adds.
After affinity studies with relevant proteins, the team extracts a mixture of the best-binding candidates and liberates the peptide tags under orthogonal oxidative conditions. The different tags are then separated and run through a mass spectrometer. ‘Then, there’s software that transforms the spectra directly into a peptide sequence,’ she adds. ‘Additionally, we also developed a Python programme that processes the data, reads the resulting sequence and identifies the Smiles string of the corresponding chemical components in the library, like a digital dictionary,’ says Rössler.
Information capacity is another key advantage of peptides – with four letters, the DNA alphabet is rather limited. The MIT team used strings of eight encoding amino acids from a pool of 16 letters, both natural and non-natural monomers. Theoretically, this hexadecimal code could convey up to 4 gigabits of data – over 4.3 billion possible chemical codes from eight distinct moieties, against a maximum of 65,000 using the same number of nucleotides. Moreover, other non-natural amino acids could further expand the chemical code, as long as they have different molecular masses. ‘We could easily double the size of our alphabet including the deuterated analogues of the amino acids used,’ says Grob.
References
SL Rössler, NM Grob et al, Science, 2023, 379, eadfl354 (DOI: 10.1126/science.adf1354)














No comments yet