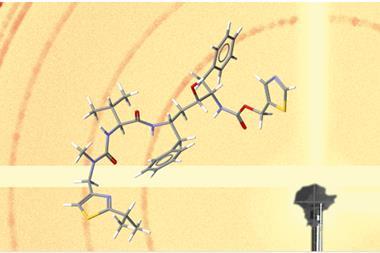
Researchers in the US have combined genome mining with a type of cryo-electron microscopy to discover new natural products and their structures – even correcting the structure of a natural product first isolated 25 years ago. The technique is faster and overcomes limitations of other methods like nuclear magnetic resonance (NMR) and x-ray crystallography, so it could boost drug and agrochemical discovery.
Natural products – compounds from living organisms – offer an abundance of potent and selective molecules that could keep up with drug resistance. Despite advances in genomics and synthetic biology, their rate of discovery has waned. This is largely because it’s difficult to determine their structures with traditional methods, such as NMR spectroscopy and x-ray diffraction, due to insufficient quantities and crystals being too small.
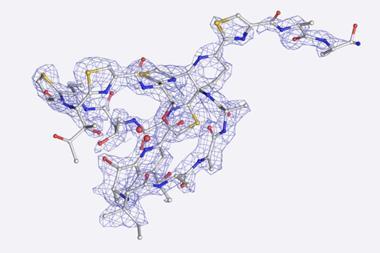
In 2018, Hosea Nelson at the University of California, Los Angeles, and his colleagues adopted microcrystal electron diffraction (microED), a type of cryo-electron microscopy (cryo-EM) – the 2017 Nobel prize-winning technique for biomolecule imaging. MicroED was originally developed in 2013 to determine protein structures, but Nelson showed it could work on tiny amounts of nanocrytalline material by shooting an electron beam through samples to collect diffraction patterns. Compared with regular transmission electron microscopy (TEM), microED works at much smaller resolutions – TEM is limited to around 0.2nm.
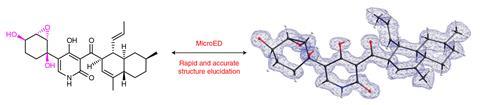
Now, Nelson’s group has used microED in conjunction with genome mining – scouring genome databases with computers to predict promising natural compounds – to accelerate natural product discovery and their structural elucidation. While this can take weeks or months with other methods, Nelson’s team discovered a new natural product from a Penicillium fungus and obtained its structure within two hours.
The researchers also identified genes responsible for producing fischerin, a natural product first isolated 25 years ago from another fungus. This enabled them to find its biosynthetic pathway and revise its structure, which previously had ambiguous stereochemistry.
‘This is an elegant demonstration of the power of using microED for the elucidation of natural product structure,’ comments Paul Race, who investigates natural products at the University of Bristol, UK. ‘Determining the structures of complex natural products is notoriously challenging due to their often elaborate chemical architectures, which frequently contain a multitude of chiral centres,’ he adds.
‘We hope the research community can adopt this as a standard technology, just like NMR and MS [mass spectrometry], in the workflow of small molecule structural elucidation,’ says Nelson’s colleague Yi Tang.
‘I’m in little doubt that this technique will become much more widely adopted in the natural products community – it has tremendous potential to expedite the process of chemical structure elucidation,’ Race says. However, he suggests access to cryo-EM instruments for data collection could be a barrier.
Race also points out that structure elucidation is only one piece of the puzzle for natural product discovery. ‘The major benefit of the approach will only be realised when applied in combination with complimentary molecular, genetic, biochemical and computational methods.’
References
L J Kim et al, Nat. Chem. Biol., 2021, DOI: 10.1038/s41589-021-00834-2








No comments yet