Cell thrives with six nucleotides instead of the normal four and new DNA code could lead to new, unique proteins

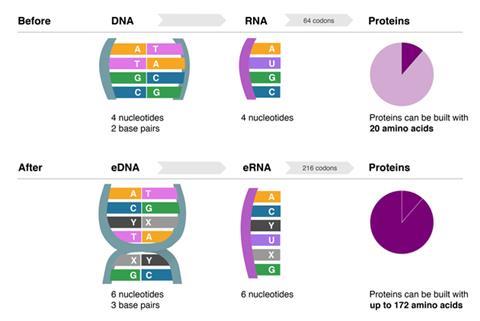
The first organism that can grow and replicate with an unnatural base pair in its DNA – giving the cell six nucleotides instead of the usual four – has been created by scientists in the US. Though efforts to engineer artificial nucleotides have been ongoing for decades, this is the first artificial base pair that a living cell can actually incorporate into its own DNA, giving it three base pairs instead of two. The technology could pave the way for advances in synthetic biology, allowing DNA to code for up to 152 new amino acids to produce unique proteins.
The advance ‘takes us closer to an expanded DNA biology that will have many exciting applications’, says lead author Floyd Romesberg from the Scripps Research Institute. Within every organism that exists in nature, DNA is made up of just two base pairs – adenine and thymine (A–T) and cytosine and guanine (C–G). But now, Romesberg and his colleagues have engineered a ‘semi-synthetic’ strain of Escherichia coli that contains an additional base pair, which they dubbed X–Y.
New base pairs
Romesberg’s lab has been trying to expand DNA’s four-letter alphabet for 20 years. Over the past decade the group have engineered several new nucleotides that have similar binding affinities to their natural counterparts, and can line up alongside natural base pairs as part of a complete string of DNA. Their best-performing artificial base pair consists of two molecules called d5SICS and dNAM that pair up using hydrophobic interactions. DNA containing these bases can be replicated in vitro by a DNA polymerase enzyme – part of the cellular machinery normally involved in DNA replication.
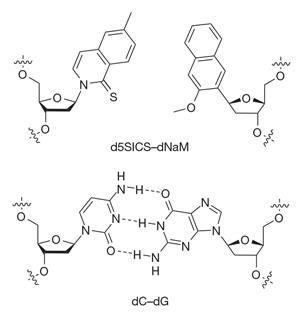
The next step, explains team member Denis Malyshev, was to integrate this base pair into a living organism – a ‘very challenging task’. The group synthesised a plasmid – a circular section of bacterial DNA – that contained a single d5SICS–dNAM base pair. This was successfully taken up by E. coli cells.
But getting the cells to replicate posed a problem. Because the new bases are not made within the cell, the team had to find a way to supply them to the bacteria, so they could be included in new copies of the DNA.
‘The key was to engineer a line of E. coli with a unique transporter protein that can specifically take up the synthetic bases,’ explains Malyshev. ‘This was necessary to make the unnatural nucleoside triphosphates – the building blocks for DNA replication – available inside the cell.’ After searching different species the team found a transport protein from the diatom algae Phaeodactylum tricornutum that could ferry the triphosphates of d5SICS and dNAM across a cell membrane. When E. coli are engineered to express this protein they can take up artificial bases added to the growth medium. This system makes replication of the new base pair controllable – if the artificial bases are no longer ‘fed’ to the cells, they continue to replicate substituting the X–Y base pair with a natural C–G pair.
After letting their semi-synthetic cells grow and divide normally, the team used sequencing techniques to show that the new base pair was retained in replicated plasmids. Crucially, the X–Y bases were not removed by the cells’ error-correcting machinery, which normally weeds out abnormalities in DNA. After 15 hours growth, the level of retention of the unnatural base pairs was estimated at 99.4% per doubling.
Exciting applications
The breakthrough could open up numerous possibilities in synthetic biology, the team say. New base pairs could expand cells’ protein production capacity – a larger number of bases means a greater variety of amino acids can be coded for – up to 172. New base pairs could also be used to engineer new regulatory architecture in cells by including them within the promoter or repressor sections of genes and producing partner proteins that can bind to them.
‘This in vivo replication of X–Y into DNA allows for the increased storage of information, which will improve the discovery and development of new drugs, diagnostics and vaccines, as well as creating innovative products, including research reagents, aptamers and nanomaterials,’ says Malyshev. The group have formed a spinout company – Synthorx – that will explore some of these possibilities.
The next step will be to see whether the new base pair can be transcribed into RNA. It is early days, says Steven Benner of the Foundation for Applied Molecular Evolution, who also works on artificial bases. ‘Transport of [nucleoside] triphosphates from the outside of the cell to the inside avoids the difficult problem of building a cell with a new metabolism that makes unnatural nucleoside triphosphates from scratch,’ he says. But he explains that this can also be a limitation. ‘Because the system requires that triphosphates be put into the “food” that the cell eats, it will have few commercial applications in its current form.’
Nevertheless, Benner praises the group’s work as a ‘major accomplishment’. ‘For 30 years, the field of synthetic biology has climbed a “wall of doubt”. Most molecular biologists and biotechnologists have doubted that any of this would work, especially the replication of DNA containing unnatural building blocks inside a living cell,’ he says. ‘Accordingly, progress has been made slowly by bold researchers like Floyd [Romesberg], who have chipped away at this wall, one experiment at a time.’








No comments yet