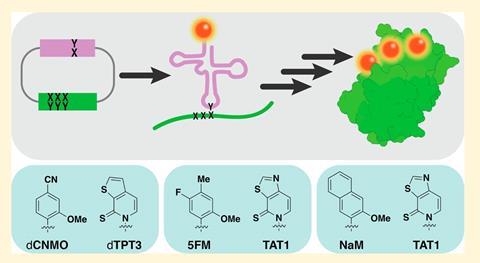
Researchers have expanded life’s genetic alphabet with six unnatural DNA base pairs, and shown that adding one artificial base pair into a bacterium can double the amount of genetic information it can store. Semi-synthetic organisms can use the additional letters just like natural ones and translate them into proteins not found in nature – a technology that could lead to tailor-made protein drugs.
DNA molecules contain four naturally occurring nucleotide bases – A, T, C and G – which partner up to form two complementary pairs. For over 25 years, researchers have attempted to expand the DNA alphabet by seeking additional nucleotides that pair up to form unnatural base pairs (UBPs).
Two years ago, Floyd Romesberg’s lab at the Scripps Research Institute in La Jolla, US, engineered UBPs into the DNA of a living organism, the E. coli bacterium, for the first time. The resulting semi-synthetic organism could produce unnatural proteins containing amino acids beyond the usual 20 that are encoded for by A, T, C and G.
Now, Romesberg’s group has synthesised more than 150 unnatural nucleotides, and evaluated a library of thousands of possible base pairings in living bacteria. In doing so, the team has identified six functioning UBPs, the best of which enables semi-synthetic organisms to store more than double the amount of genetic information that ordinary DNA does.
‘It’s desirable to have semi-synthetic organisms with extended genetic alphabets because it allows you to make proteins built from amino acids that normal proteins cannot be built from,’ explains Romesberg.
The work also bolsters the idea that the chemistry of life isn’t as unique as once thought and that potential extraterrestrial lifeforms could have different ways to store biological information. ‘Because our UBPs pair via hydrophobic and packing forces, which are completely different from the complementary hydrogen-bonding forces that nature uses, I think the results suggest that not only are the molecules that life uses not unique, but the that forces that nature uses are not the only ones that could be used,’ says Romesberg.
The synthetic nucleoside triphosphates are fed into the bacteria using a transporter protein that was previously identified from a species of algae. Once the triphosphates are in the cell, they are readily accepted and used by the organism’s normal machinery to store genetic information and make unnatural proteins.
‘Natural biological systems have not evolved to accept molecules that they have not previously seen,’ says Steven Benner, a pioneer of expanding the genetic alphabet at the Foundation for Applied Molecular Evolution, Alachua, US. ‘The Romesberg group has done a masterful intensive job exploring the space to find pairs that natural systems tolerate.’
‘The next level of challenges are relatively clear-cut,’ Benner continues. ‘We still need to engineer living cells so that they make the triphosphates of nucleosides in their own engineered metabolism. Feeding triphosphates in the growth medium is rather expensive.’
‘I would like to create a whole ecosystem of semi-synthetic life, including other bacterial semi-synthetic organisms. Maybe we could even build multi-cellular semi-synthetic organisms,’ says Romesberg. ‘Ultimately, I would like to see the platform emerge as a method to make new drugs that have a real, practical impact.’
References
A W Feldman et al, J. Am. Chem. Soc., 2019, 141, 10644 (DOI: 10.1021/jacs.9b02075)






No comments yet