Two bacterial enzymes can break up PET polymer into its building blocks
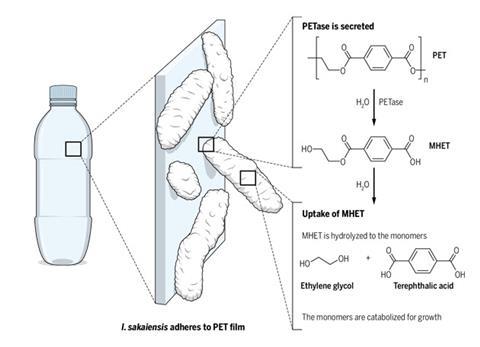
Sifting through debris at a plastic bottle recycling plant has led to the unearthing of a plastic-munching microorganism that can break down polyethylene terephthalate (PET). The researchers who discovered the bacterium hope that it will provide a new way to recycle PET plastics by breaking them down into their building blocks.
Found in everything from plastic bottles to clothing, PET fibres are produced from crude oil and lauded for their moisture resistance, making the material ideal for preserving food. Estimates suggest 50 million tonnes are produced globally every year. By 2025, this number could near 100 million tonnes. But as demand for the material grows, so do the mountains of plastic waste in landfills, as PET-based products are notoriously difficult to recycle.
To tackle this problem, Shosuke Yoshida from the Kyoto Institute of Technology and his colleagues wanted to find out whether a bacterium existed that used PET as a carbon source. But to find such a species is like looking for a needle in a haystack.
‘The environmental samples, [including] sediment, soil, wastewater and activated sludge, were collected at a yard of a PET bottle-recycling factory in Sakai city, Osaka,’ says Yoshida. The team collected 250 PET-contaminated samples in total, cultivating 1g of each in test tubes using yeast extract, ammonium sulfate, a phosphate buffer and a modified lettuce and egg medium.
By placing a small, 0.2mm-thick PET film into each medium, Yoshida was able to look for microbial cultures that thrived on the polymer. One sediment sample stood out. After 15 days, an evolving microbe culture had eaten through the PET film, converting 75% of the PET carbon into carbon dioxide.
The team isolated the bacterium that metabolises the plastic film – Ideonella sakaiensis, a Gram-negative proteobacteria – and identified which enzymes aid its digestion. ‘We revealed the genomic DNA, and searched genes that are likely to be involved in PET hydrolysis,’ explains Yoshida. ‘We were interested in one gene that is similar to a hydrolase that can hydrolyse PET.’
After finding the gene, the team cloned it and expressed its protein, PETase, finding it hydrolysed the polymer chain into mono(2-hydroxyethyl) terephthalic acid (MHET) subunits. But the bacterium doesn’t stop at MHET. Yoshida explains a second enzyme, MHETase, breaks apart MHET into the monomers ethylene glycol and terephthalic acid. ‘These monomers will be completely metabolised by this bacterium to obtain energy and cell components,’ comments Yoshida.
In reducing PET to its basic building blocks, the bacterium doesn’t just offer a way to remove PET from the environment, it could be a seed for recycling it – a possibility not lost on Yoshida. ‘In bottle-to-bottle chemical recycling, chemically-broken down PET [is] re-synthesised to make new PET,’ he says. ‘It would be possible to replace this breakdown process with [a biological one].’
Georg Guebitz, head of the institute of environmental biotechnology at the University of Natural Resources and Life Sciences, Austria, agrees the research may offer an advantage over existing techniques. ‘In contrast to other recycling methods, due to the high specificity of the enzymes, PET building blocks could even be extracted from mixed plastics waste or from composite materials.’
References
S Yoshida et al, Science, 2016, DOI: 10.1126/science.aad6359










No comments yet