Mapping out the chemical space of peptide antibiotics offers an efficient way to find new compounds
Researchers in Switzerland and Italy have devised a way to chart protein-based antibiotics according to their chemistry. This map of the chemical space has allowed them to search for new compounds more intelligently and has already led to them finding a new antibiotic for a highly resistant hospital bug.
In the biochemical arms race between bacteria and medicine, novelty is key. New types of molecules, acting in new ways, can kill microbes that are resistant to our existing arsenal. Unfortunately, the world of potential molecules is huge and mostly uncharted. New antibiotics act as landmarks, signposting where other useful compounds might lie. Researchers then start exploring nearby – although in an abstract chemical space, ‘nearby’ can be a tricky concept.
Small proteins are a rich source of new antibiotics called antimicrobial peptides (AMPs). Many of these AMPs derive from natural molecules, but Jean-Louis Reymond of the University of Bern and his colleagues have developed a potent new class of bicyclic AMPs that don’t exist in nature. They faced the challenge of finding new variations on these molecules, but the chemistry involved made an unguided search impractical. ‘We would have had to invest probably two additional years: one year to optimise the synthesis for combinatorial chemistry and a second to perform the screening and find out if our hits were real,’ explains Reymond.
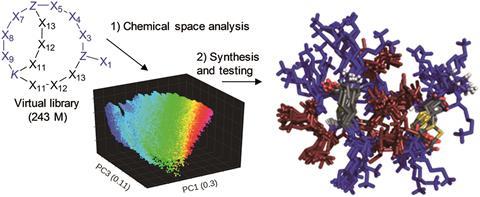
To search more efficiently, the team needed to understand how hundreds of thousands of bicyclic AMPs related to one another. They created a molecular fingerprint that digests down information on where the amino acids sit in the peptide, turning it into a set of coordinates in a chemical space. ‘The power in this approach is that the coding of chemical space is built into peptide space,’ notes Lee Cronin from the University of Glasgow, UK, whose interests include digitising chemistry. ‘This allows a new type of search to be done exploring the topological and conformational space, and I think this is significant.’ By plotting their new peptides in this space, they could intuitively understand which molecules were most related and most different.
Now that they were able to navigate in chemical space, the team took a new bicyclic peptide and devised a catalogue of related structures. However, instead of making and testing the whole library, they focused their attention – and lab time – on a representative sample that covered the space occupied by the AMPs. Once they had identified the best AMPs, the team explored near those molecules for related but better antibiotics. The researchers then applied their biochemical expertise to refine the very best molecules and understand why they were so effective.
The end product is a set of new AMPs that kill Pseudomonas aeruginosa, a tenacious, biofilm-constructing, antibiotic-resistant pathogen often responsible for hospital infections. Reymond notes that the resulting drugs are simple to produce and stable in the body, and that their novel structures mean that the pathogen doesn’t show any resistance.
With the revival of peptides and growth in the use of large molecules as drugs, Reymond expects that these approaches will become more important. ‘In this field of peptides not much has been made in terms of computational approaches; it’s time to apply all that is known for small molecules to these larger ones, and in fact one must come up with substantially new ideas to go at these larger molecules.’ Cronin sees great potential in these methods: ‘In the future I could see that this could be expanded using machine learning training algorithms … in fact we are doing this in my lab, so I am very excited that this type of data is becoming available.’







No comments yet