First DNA-based roller is 1000 times faster than current DNA motors
DNA machines that walk are fast becoming popular in the nanotechnology community, but their leisurely pace and fragility has left them in the slow lane. Now, scientists in the US have developed a DNA roller that is 1000 times faster than most synthetic DNA-based motors and can also pinpoint single mutations.
Since Ned Seeman first came up with the concept of DNA nanotechnology in 1982, the field has created a range of engineered structures that can fold into exotic shapes or walk along a linear track. Such DNA walkers generally have two legs and bind to a surface to create DNA complexes. The complexes are hydrolysed by a catalytic oligonucleotide, and the legs start taking steps.
But for some researchers, DNA walkers may be trying to run before they can walk. ‘The problem with generating a motor or machine that moves using DNA so far has been the trade-off between the speed and the fidelity,’ says Khalid Salaita from Emory University. ‘A motor, for example, that has two legs will travel fairly fast, but the problem is that Brownian motion and entropy is quite high for a two-legged machine, and these will tend to dissociate and fall off the track.’
Salaita explains that one way to get around this is to give the machine more legs, but the cost will always be speed. To address this problem, the team wanted to create a DNA roller that could circumvent the trade-off completely.
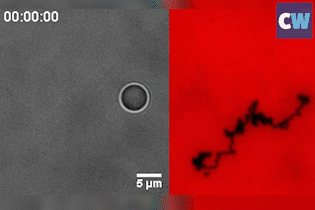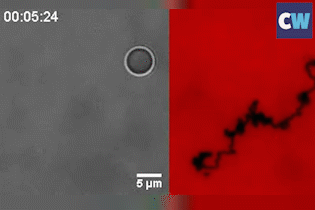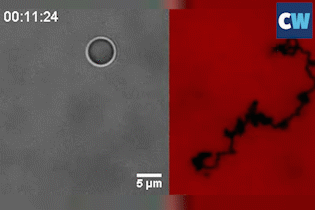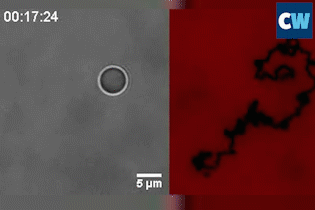
Salaita and his colleagues coated a silica bead with single-stranded DNA, before placing the ball on an RNA-coated gold surface. Up to 1000 DNA–RNA duplexes form between the ball and surface and keep the roller anchored in place. The researchers then added a biological enzyme, RNase H, to the system, which selectively hydrolyses and destroys double-stranded RNA.
They see me rollin’
Known as a ‘burnt-bridge’ mechanism, the DNA will subsequently look for new RNA to bind to. This simple reaction is what causes the ball to roll, with an average velocity of up to 1.9µm per minute. It’s so effective that the team were able to flip the sample upside-down and the ball still rolls at the same speed – inertia or gravity plays no part.
It’s the first example of doing ballistic or linear motion without an external field or force
Although such a roller will move in a self-avoiding and random manner, never crossing its former path, it can also travel in a straight line when two balls are joined together. ‘It’s the first example of doing ballistic or linear motion without an external field [or] force,’ Salaita says.
The roller is also capable of detecting single base mutations. ‘We saw that they … have specific velocities and it turns out the speed of the motor depends on the rate of DNA and RNA binding, and the rate of the enzyme destroying the RNA,’ he adds. ‘If you have a single base mutation in the RNA, that leads to a change in the velocity of the motor.’
All of this was not detected on advanced microscopy equipment, however; the team watched the roller by gluing the plastic lens of a laser pointer to a smartphone camera. Salaita hopes such a demonstration will prove DNA nanotechnology’s worth in diagnostics for ‘resource-limited settings’.
Jong Hyun Choi, a nanotechnology engineer from Purdue University, US, is impressed by the design and believes it will be a welcome addition to a growing family of DNA motors. ‘This is definitely an interesting study,’ he tells Chemistry World. ‘This new approach will be added to the array of walker designs, operating at a much larger length scale.’
References
K Yehl et al, Nat. Nanotechnol., 2015, DOI: 10.1038/nnano/2015.259











No comments yet