An optical-microscopy technique has been created that can distinguish between tiny objects less than 1nm apart. The technique could enable living cells to be viewed in greater detail than ever before.
The new method, dubbed resolution enhancement by sequential imaging (Resi), means the resolving power of fluorescent microscopy can now operate in the Ångström range for the first time. Conventional super-resolution microscopy can distinguish between objects between 15 to 20nm apart, but this does not directly translate to experiments in cells. When super-resolution microscopy came along it was so revolutionary in the life sciences that its developers won the chemistry Nobel prize in 2013.
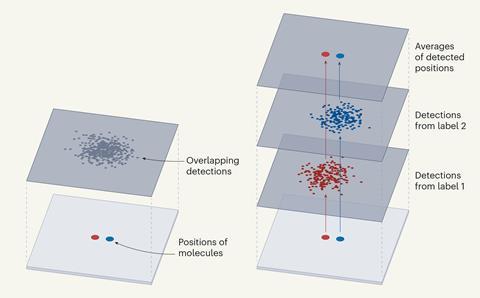
In contrast to existing techniques, Resi involves labelling adjacent copies of the same target molecule with a different tag. The researchers showed that when each of the tags was lit up, they could be clearly differentiated, even if closely spaced together, by imaging them sequentially.
By collecting multiple signals from each tagged molecule, the average position of each set could be determined with a higher level of precision than is possible with a single tagged molecule, to obtain a ‘super-super resolution’ image.
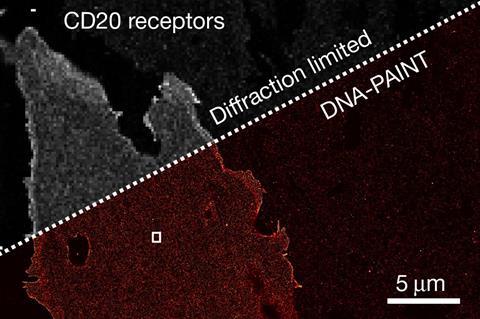
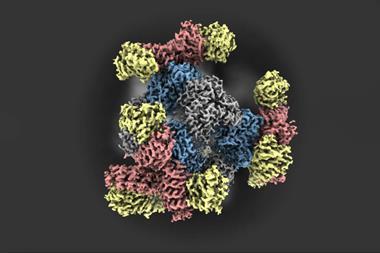
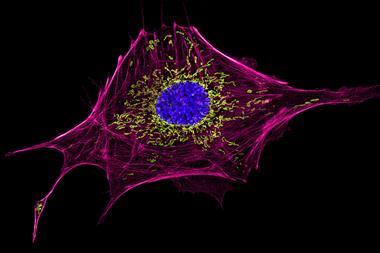
Using this technique, the researchers showed that Resi could distinguish between two tags just a few atoms, or less than a nanometre, apart in an engineered structure made of DNA. They were also able to label and image CD20, a protein targeted by the anticancer drug rituximab. By mapping the molecular arrangement of CD20 in situ in untreated and drug-treated cells, Resi was able to reveal new information about how the protein is arranged and how that arrangement is affected by binding by the drug.
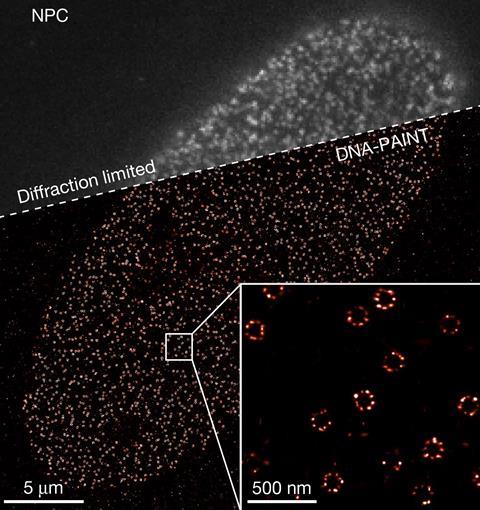
Using Resi, the researchers were able to resolve more than 2000 DNA origami structures and, separately, 1000 nuclear pore complex proteins in areas of around 67µm2 in 100 minutes. This, they added, made the technique applicable as a ‘sufficiently high-throughput tool’ for cell biology.
However, a limitation with the method was that structures observed by Resi must stay still long enough to gather precise information. For some biological experiments this means that cells must be ‘fixed’, which can introduce structural distortion and prevent dynamic processes from being visualised over time.
The size of the tag is also a limiting factor because it is the position of the fluorescent part of a tag that is observed, rather than the molecule of interest that the tag is connected to.
Despite this, the researchers said that Resi could be used as a pre-screening method for personalised treatments and could be a tool in drug design.
‘Resi is … poised to close the gap between 3D fluorescence super-resolution microscopy in whole intact cells and cryo-EM structural studies of individual supramolecular complexes, introducing a paradigm shift in fluorescence imaging by pushing optical microscopy to Ångström resolutions.’
‘We are very excited about the development of Resi, which – for the first time – allows researchers to resolve sub-nanometre distances using off-the-shelf instrumentation,’ says Ralf Jungmann, a group leader at the Max Planck Institute of Biochemistry and one of the authors of the study. ‘It is a paradigm shift for optical microscopy, enabling intramolecular imaging and closing the gap between super-resolution microscopy and structural biology studies. Resi is poised to deliver information key to understanding complex biological systems.’
Xiaolin Nan, assistant professor of biomedical engineering at Oregon Health and Science University, tells Chemistry World that Resi is ‘very exciting’. ‘I believe the method represents a major advance in biological imaging with significant implications in many areas of bioscience including structural biology,’ he adds.
References
SCM Reinhardt et al, Nature, 2023, DOI: 10.1038/s41586-023-05925-9













No comments yet