Scientists have simplified a process that harnesses the promiscuity of post-translation enzymes for making and enhancing complex molecules.1 The team behind the work say it could be developed into a platform technology for generating libraries of compounds with the structural diversity of natural products that are also new to nature.
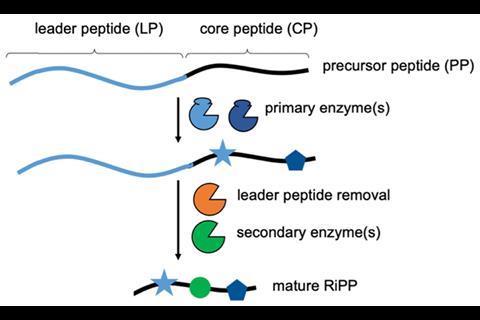
Ribosomally synthesised and post-translationally modified peptides, or RiPPs, are a class of natural products with broad structural diversity. RiPP biosynthesis begins with the ribosome translating a structural gene to generate a precursor peptide. Enzymes then modify this in multiple steps to produce the final biologically active product. ‘The way RiPP enzymes work is quite interesting; it’s different from how enzymes usually work. The enzymes bind to one bit but do catalysis somewhere else… the result of that is that a lot of these enzymes are highly promiscuous,’ explains Jesko Koehnke from the University of Glasgow, UK.
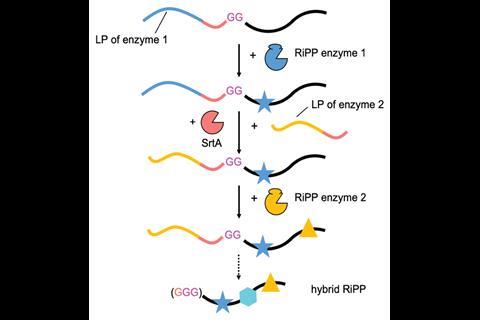
Although it could potentially be used to create diverse libraries of compounds, at present, RiPP engineering is limited by the need for a custom-designed precursor peptide with a leader section that the post-translation enzymes can bind to. In many cases, the peptides must also contain a specific sequence of amino acid residues so enzymes recognise them as a substrate – the enzyme will then modify a different part of the peptide known as the core peptide. Resulting compound libraries are currently variations on a theme, as determined by the chemistry of the enzyme used. Koehnke wanted to explore if more diverse libraries would be possible if such dependence on the leader peptide were eliminated.
‘We tried different ways and it wouldn’t work; we tried to do it chemically and with other enzymes. In the end, I went to a series of talks and there was a guy [Dirk Schwarzer at the University of Tübingen in Germany] who talked about sortase A and then all of a sudden it was easy,’ recalls Koehnke. Sortase A is a bacterial enzyme that cross-links peptidoglycan chains, which structural biologists have adopted for fusing proteins together. Koehnke and Laura Franz of Saarland University in Germany have now shown that sortase A can be used to swap leader peptides between biosynthetic steps, making it much easier to instruct a variety of enzymes to modify ribosomal products.
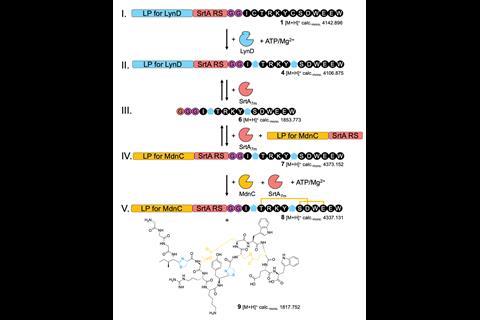
As a proof-of-concept for their leader peptide exchange technique, Koehnke and Franz combined enzymes from two unrelated biosynthetic pathways originating in different cyanobacteria. They began by modifying their precursor peptide with LynD, an enzyme that converts cysteine residues into thiazoline heterocycles. The precursor peptide contained a leader section for LynD to bind to, followed by the recognition motif of sortase A and was joined to the core section of the peptide by two glycines. They then introduced sortase A, which cleaved two of the residues in the motif, before catalysing a new amide bond between this end of the peptide and the leader peptide for the next modifying enzyme, MdnC. MdnC catalysed ester bond formation between the side chains of certain residues. A final step reintroduced sortase A to liberate the desired hybrid product.
In 2017, scientists from the University of Illinois at Urbana−Champaign reported a method for making RiPP hybrid products using a chimeric leader peptide with recognition sequences for two different enzymes, so that two enzyme modification steps could occur sequentially.2 This was a big step forward in making hybrid natural products, but users needed to know the recognition sequences associated with each enzyme. Koehnke and Franz’s method is less involved; they say it has the potential to be developed into a plug and play approach in which compatible enzymatic steps can yield complex compounds that would be unfeasible to make using conventional synthetic chemistry.
‘RiPP natural products are a very exciting class of potential drugs to address everything from antibiotic resistance to cancer,’ notes Michael Jewett, whose laboratory at Northwestern University in the US also works on ribosomal synthesis. ‘Thus, this approach may enable novel compound libraries to be built, which were previously inaccessible, to identify unique biotherapeutic activities.’
References
These articles are open access
1 Laura Franz and Jesko Koehnke, Chem. Commun., 2021, DOI: 10.1039/d0cc06889f
2 B J Burkhart et al, ACS Cent. Sci., 2017, 3, 629 (DOI: 10.1021/acscentsci.7b00141)








No comments yet