Chemists in the UK and Belgium have made molecular machines that emulate how cells naturally produce proteins, but read their instructions from threads based on characteristically-unnatural polystyrene. The group led by David Leigh from the University of Manchester use this ‘cyborg ribosome’ to make a simple protein that coils up into an asymmetric catalyst. That catalyst itself then guides an epoxidation reaction to selectively make one of two possible enantiomer products.
‘Biology is based on just four sequence polymers: DNA, RNA, proteins and carbohydrates,’ Leigh says. Molecular machines that integrate other polymers might impart entirely new properties to them, like those seen in biology, he adds. ‘Perhaps we could have polystyrene polymers that could be muscles or as strong as spider silk.’
Biology has had one and a half billion years to perfect the ribosome, so give us a bit of time
David Leigh, University of Manchester
Leigh’s team had previously developed similar systems to mimic ribosomes, the molecular machines in our cells that read genetic information about which amino acids to bolt together. Then, they threaded a large ring-shaped rotaxane molecule around a protein ‘track’. The rotaxane could move down this track, bolting together three amino acids. ‘The track in that case was quite complicated to make,’ Leigh explains. Looking to broaden the potential of rotaxane-based machines, Leigh and his team thought ‘it would be much more interesting to have these things working on conventional polymers’.
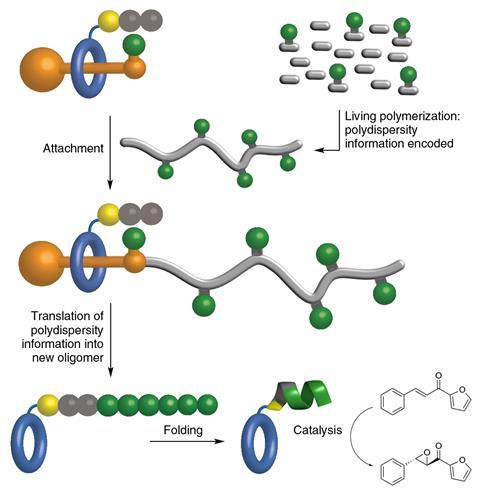
However, making that switch posed a potential challenge in controlling which amino acids to add, and how many. Ribosomes decode instructions from codon groupings arising from bunching the four letters in the RNA code together in threes. By contrast, polystyrene threads repeat monomers in similar patterns for long stretches.
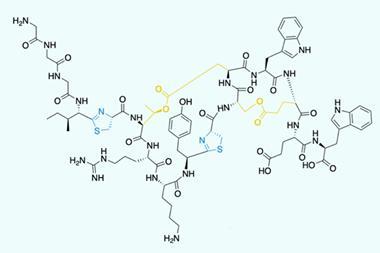
Leigh’s team targeted a protein with just one type of amino acid, leucine, because leucine chains long enough to coil up are already known to be asymmetric epoxidation catalysts. The scientists realised that making the polystyrene thread that fed into the rotaxane the right length would in turn ensure the leucine chains were long enough to do this.
The chemists therefore used atom transfer radical polymerisation, which gives polymer chains with a relatively narrow range of lengths compared to other techniques. In this way, they combined normal styrene monomers with some that had leucine attached, giving threads with 52 normal styrene monomers and six monomers with leucine, on average.
Firing up molecular machines
Driving the rotaxane along the thread over four days gave leucine chains that the team then used to catalyse the epoxidation of chalcone molecules. After some troubleshooting, they were able to claim that the leucine helix enabled 100% conversion. The reactions the chemists used it in made 96% of the desired enantiomer, and 4% of the other possible product, albeit only on the milligram scale.
This is as active and selective as similar previously-described catalysts, points out Anne-Sophie Duwez from the University of Liége in Belgium. Such molecular machines’ ‘use in concrete applications may be a long way off, but the results described here show that it is possible to design and synthesise a system with useful properties’, she says. ‘Any organic chemist would be delighted with a catalyst with 100% conversion efficiency and 92% enantiomeric excess. Of course the challenge remains the synthesis of the machine itself.’
Stephen Goldup from the University of Southampton, UK, calls the approach ‘really elegant’, and echoes Duwez’s point about the challenge making such machines currently poses. However, ‘to worry about this is to miss the point’, he adds. ‘It’s only in the last few years that artificial molecular machines have even begun to perform such challenging tasks and their sophistication is increasing rapidly,’ Goldup says. ‘It’s hard to predict what the ultimate outcome of this new direction will be but given the central role that similar machines play in living systems, it seems likely their artificial counterparts will play a key role in making challenging molecules in future.’
Leigh concedes that this machine is crude compared with its biological inspirations, but stresses that it’s an early step in a long journey. ‘The ribosome puts together 15–20 amino acids a second. For us to take four days to put together six or seven is not that impressive yet. But biology has had one and a half billion years to perfect the ribosome, so give us a bit of time.’






No comments yet