An artificial protein moves between predetermined conformations when exposed to calcium ions. These dynamic structural changes mimic those seen in naturally occurring proteins, which often gain their functionality by switching between subtly different arrangements.
Designing useful proteins that do not occur naturally is an art that researchers have been picking away at since the 1980s, greatly accelerated in recent years with the help of machine learning algorithms. However, engineering ‘de novo’ proteins that make controlled conformational changes has been difficult to achieve.
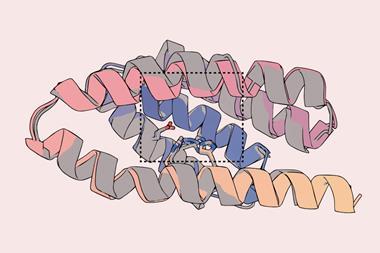
While researchers have previously made de novo proteins that can change their conformation, these changes have involved a large portion of the protein moving wholesale with respect to the rest of the protein, for instance at a hinge. Now, Tanja Kortemme, a bioengineer at the University of California in San Francisco, US, and her colleagues have designed an artificial protein that makes more subtle structural changes in a manner similar to many naturally occurring proteins.
As Kortemme points out, a lot of processes in living organisms rely on signal transmission based on small changes in protein conformation, such as an internal feature twisting or a helix changing its orientation. These subtler conformation changes could have advantages over hinge-type changes because of the molecular specificity required for all the pieces of the protein to click into the new conformation.
‘In principle, these motions are more subtle,’ Kortemme tells Chemistry World. ‘But they are much more difficult to engineer.’
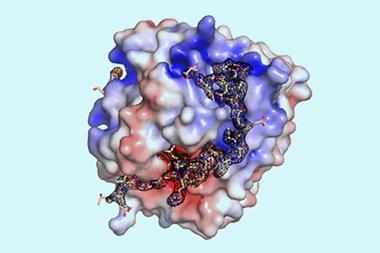
Proteins arrange their atoms into positions that minimise the potential energy of their interactions. This means that static protein design can be considered an energy minimisation problem – like finding a valley in a landscape that maps the potential energy for different configurations. Such problems become nigh on impossible using first-principle quantum calculations because there are so many atoms interacting, but machine learning tools have provided a huge helping hand in recent years. However, for switchable proteins there needs to be two arrangements where the ‘valleys’ have similar depths and are not too deep so as to make conversion between conformations unlikely. ‘You need to get the balance of the two conformations exactly right,’ notes Kortemme.
Experimental validation
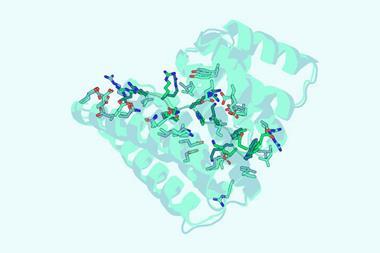
To start with the researchers essentially tweaked a naturally occurring protein – the N-terminal domain of troponin C, a protein complex involved in muscle contractions. Starting from this domain, Kortemme describes graduate student Amy Guo as the ‘intellectual driver’ in harnessing deep-learning tools to guide the design of a de novo protein that has two possible conformations, one of which would react with calcium ions to be more stable. This meant that if the protein began in the other conformation, it would switch in the presence of calcium ions.
‘To actually make the protein is one of the easiest parts,’ says Kortemme. Advances in DNA synthesis mean that they can order the genes for the proteins they have designed from suppliers simply by providing the amino acid sequence. Even the purification is reasonably straightforward, since de novo proteins tend to be quite stable and so resistant to some of the purification techniques that can destroy naturally occurring proteins.
‘Often the most difficult part of a protein design project is the experimental characterisation, especially when you’re building complex systems like these,’ says Florian Praetorius, who works on biomolecular design at the Institute of Science and Technology Austria, and who was not directly involved with the research reported here. ‘I’m very impressed at the work the Kortemme lab has done on not just designing these dynamic proteins, but also convincingly showing that they actually work as intended,’ he adds.
Kortemme explains that her team is now working on proteins devised completely from scratch, as opposed to derived from a naturally occurring protein. The researchers are also looking at ways to control how the protein moves in terms of its shifts and twists.
References
A B Guo et al, Science, 2025, DOI: 10.1126/science.adr7094












No comments yet