The science behind the prize-winning gene editing tool that could change our lives
Emmanuelle Charpentier and Jennifer Doudna have scooped the 2020 Nobel prize in chemistry ‘for the development of a method for genome editing’. Specifically, they’ve been awarded the prize for their discovery of the Crispr–Cas9 genome editing technique that allows scientists to make precise alterations to the genetic code of living organisms. Crispr–Cas9 is a powerful tool that could revolutionise many aspects of our lives, from medical treatments to the way we produce food. It’s also seen its fair share of controversy in recent years. Here, we take a deeper look at these ‘genetic scissors’ and why they’ve won the Nobel prize.
Why did it win?
Since Charpentier and Doudna began investigating the Crispr–Cas9 system in 2011, the field has exploded. Due to the relative simplicity and affordability of Crispr systems, researchers around the world have been able to apply the tools to all manner of different problems. Today there are entire journals, conferences and companies dedicated to the technique.
The ability to cut any DNA molecule at a chosen site has huge potential – from treating genetic illnesses to creating disease-resistant crops. Trials have even shown how Crispr-delivered genetic modifications can spread through populations of mosquitoes and stop malaria infections – such ‘gene-drives’ offer a way to eliminate the disease altogether. And in the face of the Covid-19 pandemic, researchers have found ways to use Crispr in rapid coronavirus diagnostic tests and have also proposed using it to attack the virus’s genome.
As Claes Gustafsson, chair of the Nobel committee for chemistry, said at the award announcement, ‘There is enormous power in this genetic tool, which affects us all.’
Crispr technology has even been used to make more delicious beer.
How does it work?
The whole Crispr gene editing tool has been adapted from the immune system of bacteria. The term Crispr comes from ‘clustered regularly interspaced short palindromic repeats’, which refers to distinct genetic sequences found in the genomes of bacteria. Each Crispr sequence is transcribed into RNA sequences that will target the DNA of a virus. These sequences also include cas (Crispr-associated) genes that code for DNA-cutting Cas enzymes. Together, the guide RNA and Cas enzyme form a complex that hunts out viral DNA and chops it up.
In Crispr gene editing, scientists repurpose this system by designing a guide RNA sequence of around 20 nucleobases that matches up to a DNA sequence they wish to target in a cell’s genome. This RNA sequence is paired with the Cas9 enzyme that will cut the DNA strand at the targeted site. The whole DNA sequence coding for both these components of the Crispr-Cas9 tool can be delivered to the target cell via a plasmid.
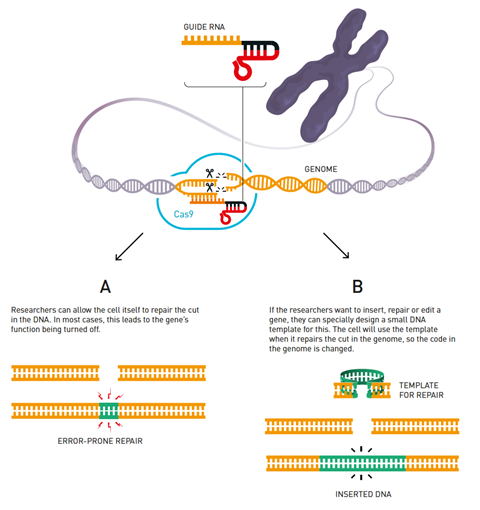
The tool can therefore be used to edit a cell’s genome with incredible precision – for example, it can cut out a dysfunctional gene associated with a hereditary illness. And if the healthy version of the gene is also delivered to the cell, the cell’s own repair system will then incorporate the healthy strands at the site where it has been cleaved.
What did the laureates do?
In 2011, when investigating the bacteria Streptococcus pyogenes, Charpentier discovered a molecule called tracrRNA that forms a key part of the Crispr–Cas system in bacteria.
Meanwhile, Doudna had been investigating the function of the cas genes, and learned that the Cas proteins they code for are involved in cutting up DNA as part of the bacterial immune system against pathogenic viruses.

That year Charpentier teamed up with Doudna to investigate the system further. Together they revealed how the Cas9 protein, CrisprRNA and tracrRNA worked together to snip DNA strands into two parts. They then simplified the system by combining the CrisprRNA and tracrRNA into a single molecule – guide RNA – making it easier to use, and showed how this could be used to cut any DNA strand at a site of their choosing, opening the door to using the tool in all manner of genome editing experiments.
While previous tools for genetic editing existed before Crispr-Cas9, the new tools are much simpler and cheaper. This has led to the huge expansion of the field by making gene editing accessible for scientists all around the globe.
So where’s the controversy?
For years Crispr has been at the centre of a long-running patent dispute. Shortly after Doudna and Charpentier’s discovery, Feng Zhang’s team the Broad Institute in Cambridge, US, patented a way to use the technique in eukaryotic cells. There have been protracted court battles between Doudna’s group at the University of California in Berkeley, US, and the Broad team over who holds the key piece of intellectual property. In the meantime, numerous groups and companies have been granted patents for many new Crispr-related technologies, meaning that as time goes on, the original patents at the centre of the dispute are becoming less relevant.
Another area of controversy surrounds the potential consequences of using genome editing tools at all. As the genome is so complex, we can’t always know what will happen when we edit genes. Some genes have multiple and often unknown functions – editing them to correct for one problem could end up creating new unforeseen ones. This is particularly important when it comes to editing germline cells (those that can be passed on to an organism’s children), because the modified genes can be inherited by future generations.
As a relatively new technique, we also know that Crispr itself isn’t perfect. Some studies have shown off-target cuts, where the tool has snipped DNA strands at additional locations to the desired site. This clearly can have harmful consequences, and so many researchers are looking into ways to improve the technique and make it more suitable for medical uses.
With these concerns in mind, scientists worldwide – including Doudna and Charpentier – have called for a moratorium on editing human germline cells, until we can know more about the consequences. Such calls intensified after the rogue Chinese scientist He Jiankui edited human embryos that were then brought to term in 2018. He is now serving a three year prison sentence for conducting the study.
What could happen next?
Several clinical trials have already begun on Crispr-based therapies, with promising reports emerging this year. In February, the first study to look at a cancer treatment using Crispr-edited immune cells reported that the modified cells were safe, with no serious side-effects in the three patients studied. While the efficacy of the treatment on the cancers was minimal, it may help to inform future Crispr-based T-cell treatments.
One month later, a patient with hereditary blindness became the first person ever to have a Crispr–Cas9 therapy directly administered into their body. And in June, the Swiss gene-editing company Crispr Therapeutics announced that two patients with beta thalassaemia and one with sickle cell disease would no longer require blood transfusions after their bone marrow stem cells were edited using Crispr techniques.
Earlier this week, Doudna launched a new company, Scribe Therapeutics, to begin work on treatments for amyotrophic lateral sclerosis.
Other Crispr-based technologies are coming closer to commercial reality. For example, the US genome engineering company eGenesis is developing ways to use the technique to edit pigs’ genes so that their organs might be transplanted safely into humans. In the agricultural sector, many companies are working on ways to use Crispr to speed up the selection process for crops with desirable traits such as disease-resistance or improved flavour.
At the fundamental level, researchers are working on ways to improve the system itself. By using alternative Cas proteins, some groups hope to make the tool more effective and easier to use in certain settings. Doudna’s group recently reported on a CasX protein that is smaller than Cas9 and potentially easier to introduce into target cells.
Additional information
Delivering DNA into cells and tissues is an important part of gene therapy, even more so for Crispr-Cas9 approaches because plasmids carrying this system are very large. This research paper describes a non-viral vector for delivering plasmid DNA carrying Crispr-Cas9 into tumour spheroids, which are good in vitro models for tissues but also challenging transfecting targets.
1 S J Zamolo, T Darbre and J-L Reymond, Transfecting tissue models with CRISPR/Cas9 plasmid DNA using peptide dendrimers, Chem. Commun., 2020, DOI: 10.1039/d0cc04750c
Regulating the function of Crispr-Cas9 is on the agenda for many researchers because the ability to restrict it in a spatial and temporal manner opens the door to precisely manipulating genomes and minimising any side effects. By introducing photolabile groups into the system, these researchers have shown how they can regulate Cas9 activity with light
2 Y Wang et al, Photocontrol of CRISPR/Cas9 function by site-specific chemical modification of guide RNA,Chem. Sci., 2020, DOI: 10.1039/d0sc04343e
It seems that Crispr-Cas systems aren’t just handy for gene editing. This paper describes how the Crispr-Cas system was used to assemble a multi-enzyme cascade containing five distinct enzymes. The team behind the work hope it could be the beginnings of a general method for building complex scaffolded biocatalytic pathways
3 S Lim et al, CRISPR/Cas-directed programmable assembly of multi-enzyme complexes, Chem. Commun., 2020, 56, 4950 (DOI: 10.1039/d0cc01174f)
And to round things off, here are some reviews on how Crispr-Cas9 works, the delivery processes for therapeutic nanoparticles and the physiological obstacles for those process
4 Y Xu, R Liu and Z Dai, Key considerations in designing CRISPR/Cas9-carrying nanoparticles for therapeutic genome editing, Nanoscale, 2020, DOI: 10.1039/d0nr05452f
5 Y Gong et al,Lipid and polymer mediated CRISPR/Cas9 gene editing, J. Mater. Chem. B, 2020,8, 4369 (DOI: 10.1039/d0tb00207k)









No comments yet