Scientists encourage soil microorganisms to express themselves

A new approach using a combination of genetic tools to capture, manipulate and express a previously silent gene cluster has led to scientists in the UK to discover a new natural product that shows promising antibacterial and anti-cancer activity.
‘Bacteria have the ability to produce an untapped reservoir of metabolites, however their production is tightly controlled by transcriptional regulators – DNA binding proteins which determine when expression is on or off,’ explains Christophe Corre from Warwick University. ‘Typically, these antibiotic-like molecules are not produced when bacteria are grown in a lab as they do not need these molecules to outcompete other microoganisms,’ he adds.
Now, Corre’s team has found a way to discover the natural products that soil microorganisms encrypt in their DNA. It involved capturing a set of silent genes that encode bioactive molecules and turning on their production. ‘[Our study] has evidenced that breaking locks at the transcriptional level does trigger the production of truly novel bioactive substances.’
The concept, using a combination of genetic tools including yeast transformation-associated recombination and clustered regularly interspaced short palindromic repeats (Crispr)-associated gene editing technologies, will work on many biosynthetic gene clusters that have already been sequenced. Corre says they do not need to sequence new genomes as ‘there is already an overwhelming number of unexploited metabolic pathways available’. This is an advantage over the Crispr-Cas9 systems currently available for editing the genome of Streptomyces bacteria, as they need to be tested from scratch for each new bacterial species.
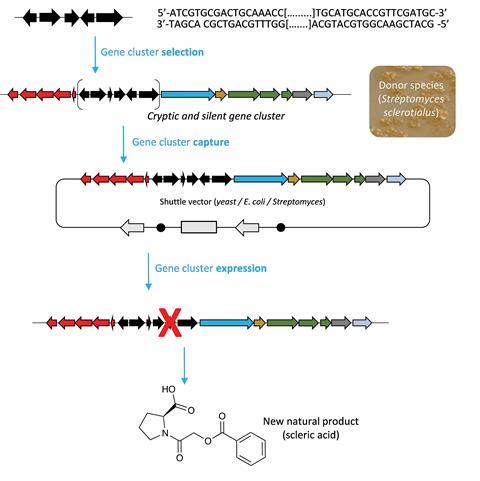
Validation of this approach has come with the discovery of scleric acid – a specialised metabolite and an unprecedented hybrid natural product. Some unusual enzymes were involved in the biosynthesis of scleric acid due to its remarkable structural features, in particular an enzyme from the ATP-grasp family that activates benzoic acid. ‘Biosynthetic enzymes catalysing scleric acid formation could be exploited as novel biocatalysts to replace chemical process, and the potential of exploiting biosynthetic enzymes as biocatalysts has recently been emphasised by Frances Arnold winning the Nobel prize in chemistry,’ says Corre.
It is difficult to predict how effective any given molecule will be against a specific target so it is common to screen any structurally new natural products against as many targets as possible. Scleric acid has already shown antibacterial activity against Mycobacterium tuberculosis as well as inhibiting an enzyme associated with cancer – nicotinamide N-methyltransferase.
‘Bacteria have biosynthetic machinery to produce a vast array of bioactive compounds, however, the majority of this chemical diversity is inaccessible,’ says Jason Micklefield, a chemical biology expert at The University of Manchester, UK. ‘This type of approach can open the way to the discovery of new bioactives for therapeutic, agrochemical and other applications.’
References
This article is open access
F Alberti et al, Chem. Sci., 2018, 2019, DOI: 10.1039/c8sc03814g













No comments yet