Today, the 2018 chemistry Nobel prize was awarded for putting the power of evolution into chemists’ hands. Frances Arnold was recognised for inventing directed enzyme evolution, while George Smith and Gregory Winter received the prize for discovering how to get bacteria to make proteins to order.
What is directed evolution and what does it have to do with chemistry?
Arnold herself summed it up nicely: ‘In directed evolution we provide a new niche in the laboratory, so to speak, and encourage evolution of enzymes to catalyse commercially useful reactions.’
Enzymes can speed up reactions dramatically while carrying out their work in water at room temperature. They are also really good at making one specific bond – without messing around with other functional groups – and are often enantioselective too. So it’s easy to understand why chemists like using enzymes to catalyse reactions.
However, many bonds chemists are interested in aren’t made by any natural enzyme. This is simply because organisms have never needed to evolve the ability to make, for example, carbon–silicon bonds.
How does it work? Can’t we just make new enzymes from scratch?
Although most enzymes are built from only 20 different amino acids, thousands of these protein building blocks can make up each enzyme. Apart from the synthesis challenge, knowing how to connect them to create a catalyst that will do something useful is incredibly difficult. This is why Arnold decided to abandon this approach and instead use nature’s sophisticated machinery that has even allowed bacteria to evolve to feed on synthetic compounds such as plastics.
Directed evolution starts off with an enzyme that has properties similar to the desired ones. In her earliest work, Arnold created an enzyme that cleaves peptide bonds in organic solvents. The natural protein only does this in water – in fact, organic solvents change its structure and stop it working.
Arnold then introduced random changes – mutations – into the gene that encodes the peptide-cleaving enzyme. Different versions of the mutated gene were then inserted into bacteria that started churning out many, slightly different enzymes. Arnold then selected the bacteria whose enzymes worked best in organic solvents and subjected them to further rounds of test-tube evolution.
After only three generations, an enzyme was created that worked 256 times better in organic solvents than the original type.
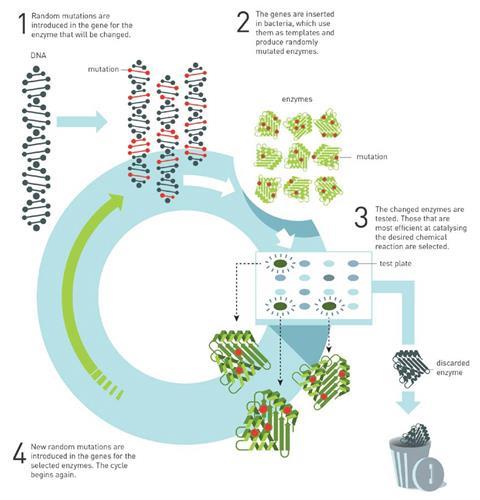
Why did this discovery win the Nobel prize?
Directed evolution had not only created catalysts that can do all kinds of complex chemistry but has also made manufacturing enzymes and biofuels more environmentally friendly.
Just this year, Arnold’s team evolved a haem protein to make strained and super strained rings. They also created a mutant Escherichia coli bacteria whose modified biosynthetic pathway converts sugars into isobutanol – a precursor for fuels and plastics.
And this work is not just an academic exercise. Many of Arnold’s creations have made it into industrial settings. Lipases, modified by directed evolution, are produced in enormous quantities and added to laundry detergents to break down fats. Arnold’s own spin-out company Provivi develops biopesticides through directed evolution.
So what about the other discovery recognised in the Nobel prize, phage display?
It uses bacteria-infecting viruses – called bacteriophages – and gets them to make any desired protein to order and then stick it to its outer shell. The method was invented by George Smith, initially as a way to find out which genes encode which proteins.
Greg Winter realised that even more could be done with this technique. He inserted genes for antibodies into phages, which promptly inserted these Y-shaped proteins into their coats. Antibodies can stick to bacteria and viruses, signalling to immune cells to destroy these invaders. They are valuable treatments for autoimmune diseases and some types of cancer.
How does it work?
Phages are extremely simple biological structures – some argue that they can’t even be called ‘alive’. They consist of some genetic material wrapped in a protein capsule. Like other viruses, they can’t reproduce themselves, but instead hijack a bacterium’s metabolism.
Smith realised that he could insert any gene into the part of the phage’s genome that contains the blueprint for its protein coat. When the phage replicates, the protein the inserted gene encodes for then ends up in the phage’s coat.
As the phages can work with almost any protein, Winter started using phages to make antibodies. To find the best antibodies for fighting disease, he exposed the antibody-carrying phages to a disease target – for example a protein or a DNA sequence.
Phages whose antibodies don’t stick well to this target get removed during a purification step. Those that perform well are subjected to a round of random mutation and the search for the antibodies that stick best to the disease target is repeated. Repeating this cycle a few times creates antibodies that strongly bind to their target and, in theory, could be used as a drug.
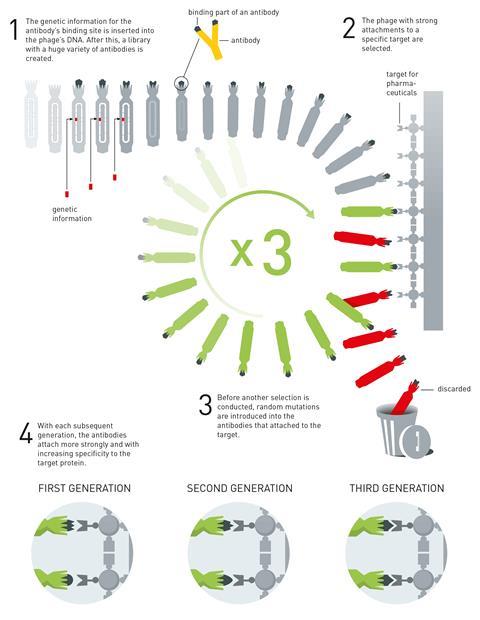
Why did phage display receive the prize?
Phage display has created an entirely new way to make antibodies. Before it was invented, scientists had to rely on mice to make antibodies, which were often not well tolerated by people’s immune systems making them poor drug candidates. The antibodies made by phages are human, so this isn’t a problem.
In 2002, the first drug made entirely by phage display was approved. Humira (adalimumab) neutralises an inflammation protein that drives autoimmune diseases such as rheumatoid arthritis and Crohn’s disease. It is now the world’s biggest selling drug and had sales of $16 billion in 2016.
But it is still early days for antibody drugs and their potential is huge. Scientists are now trying to find antibodies to kill cancer cells or treat anthrax.
This sounds like two very different pieces of research. What links them?
Phage display is essentially a different type of directed evolution. Both methods create new molecules using nature’s biosynthetic machinery: directed evolution makes new enzymes, phage display evolves better therapeutic antibodies.












4 readers' comments